Garnett Group
Translational Cancer Genomics
Research foci
There are currently four complementary research focuses in my laboratory:
- The genomics of drug sensitivity. High-throughput drug screens in human cancer cell cultures to identify genetic features of cancer cells that are predictive of drug sensitivity.
- Mapping synthetic-lethal dependencies in cancer cells. Genome-wide CRISPR-Cas9 synthetic-lethal screens in cancer cell lines to identify potential new oncology drug targets.
- A new generation of organoid cancer models. Methods for the derivation, characterisation and use of a new set of cancer organoid in vitro cell culture models.
- Tumour-immune cell interactions. The use of patient-derived in vitro co-cultures to understand mediators of tumour and immune cell interactions.
These studies are intergrated into our Cancer Dependency Map initiative, which aims to identify all dependencies in cancer cells to help guide future precision cancer medicines.
Background
Cancer is a genetic disease caused by the accumulation of changes within the DNA of cells that confers a growth and survival advantage. Cancer is not just one disease, but many different diseases, each with a different spectrum of underlying genetic causes. The functional consequence of these genetic changes in the genes of healthy and cancer cells is often poorly understood, and how they contribute to disease is often unclear. Furthermore, these genetic changes can impact on patient responses to therapy and consequently can be used to select patients most likely to benefit from a specific treatment.
The Translational Cancer Genomics team investigates how genetic alterations in cancer contribute to disease and impact on response to therapy. Our research is at the interface of cancer genomics, cell biology and cancer therapeutics and employs high-throughput biology approaches together with detailed mechanistic studies.
The team have state-of-the-art facilities to perform their research including extensive robotics, acoustic dispensing, high-content microscopy, CRISPR and chemical libraries, and access to core Sanger IT and genomics infrastructure.
Seminars and talks from the team, as well as tutorials for our public databases, are available to view on Twitter.
The genomics of drug sensitivity
The team uses high-throughput drug sensitivity screens in highly annotated human cancer cell cultures to identify genetic features of cancer cells that are predictive of drug response.
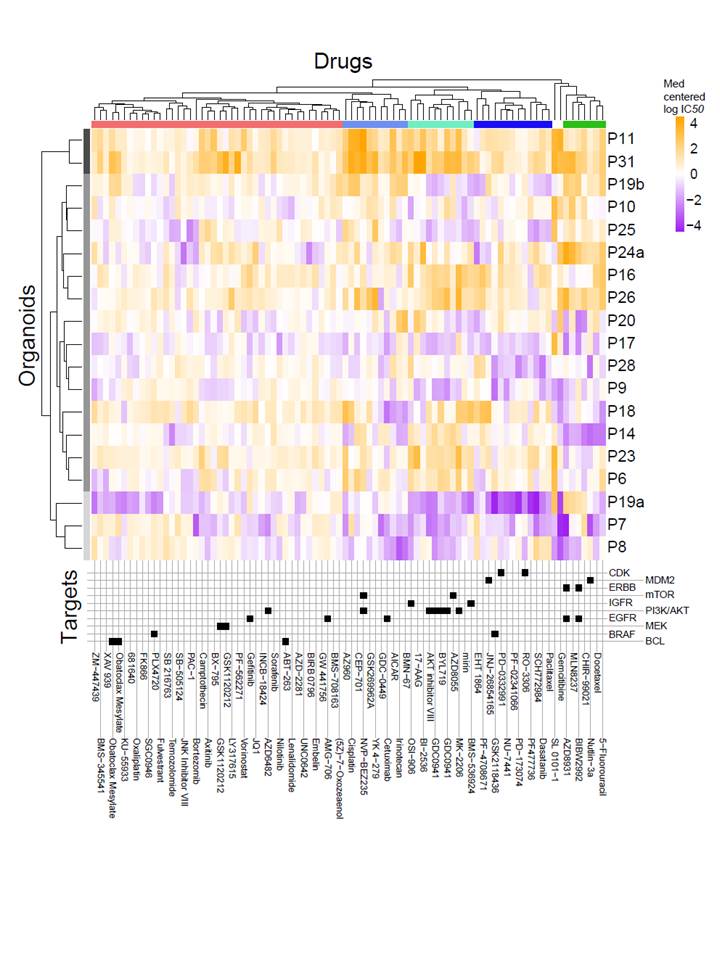
The collection of cancer cell models to study drug response includes >1000 human cancer cell lines, organoids, and engineered mouse and human cells. Cell culture models are highly annotated at the level of the genome (DNA sequencing), transcriptome (gene expression and RNAseq), epigenome (DNA methylation), and proteome. The team perform single-drug and combination screens with hundreds of anti-cancer compounds across their large collection of cell culture models to detect drug sensitivity in specific tissue and genetic sub-types.
The results from these screens are used to improve the design of clinical trials through the identification of patient populations most likely to respond to a therapy.
This research is performed in partnership with the laboratory’s of Ultan McDermott and Mike Stratton within the Cancer Genome Project at the Sanger Institute. The results of this work are available from the Genomics of Drug Sensitivity in Cancer database.
Mapping synthetic-lethal dependencies in cancer cells
Mathew’s group are performing genome-wide CRISPR-Cas9 synthetic-lethal screens in cancer cells to identify potential new oncology drug targets in defined genetic backgrounds.
The complexity and diversity of cancer genomes represents a significant challenge when developing new cancer therapies. Specifically, identifying cellular signalling nodes and processes whose perturbation selectively kills cancer cells while sparing normal cells remains acutely difficult. This is because our understanding of which proteins are necessary for cancer cell survival is incomplete. Furthermore, our understanding of cellular networks and processes is relatively poor in normal cells, let alone in the context of cancer cells with their myriad of molecular alterations. Thus, systematic and unbiased approaches to identify critical dependencies in cancer cells could significantly expand the repertoire of new drug targets for future development.
Genome-editing technologies such as CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats) are a powerful tool for studying gene function in normal and diseased cells. This approach uses a single guide RNA (sgRNA) to recruit the Cas9 endonuclease to a desired genomic loci to create double strand breaks, which are repaired through an error prone process resulting in targeted gene inactivation. Taking advantage of the programmable nature of the sgRNA, it is now possible to use a library of sgRNAs to perform genome-wide functional genetic screens across a diverse array of cellular models and systems.
Mathew’s group, in collaboration with the laboratory of Kosuke Yusa, are exploiting CRISPR-Cas9 genome-editing technology to systematically identify new drug targets using genome-wide ‘synthetic lethal’ screens in cancer cell lines. The identification of acute sensitivities, which occurs within specific molecular/genetic sub-types, could provide novel opportunities for genetically targeted therapeutic intervention. The results of our CRISPR screens and target prioritisation are available through the Project Score database.
A new generation of organoid cancer models
Approximately 1,000 human cancer cell lines are available to scientists worldwide and this has been a useful resource for cancer research. However as we enter the era of precision medicine, poor representation of some cancer types, insufficient numbers to capture the genetic diversity of cancer, lack of clinical outcome data and lack of comparison to normal reference sample limit their use.
Novel cell culturing methods such as organoid derivation have revolutionised our ability to derive cell line models from both healthy and diseased tissue, and have the potential to overcome these limitations.
We are generating and characterising new patient-derived cancer cell line models from different tumour types as experimental tools. These cell lines are being characterised at the level of the genome and transcriptome, profiled for differential sensitivity to anti-cancer therapies, and are being made available to the research community. These models are being generated as part of an international collaboration called the Human Cancer Model Initiative, led by the Sanger Institute, the U.S. National Cancer Institute, and Hubrecht Organoid Foundation.
Organoid models generated by Sanger are for sale through American Type Culture Collection.
Our Cell Model Passports database hosts cell model genetic and functional datasets.
We anticipate that this highly annotated resource will have broad applications and serve to catalyse a new wave of discovery in fundamental cancer biology and therapeutics.
Tumour-immune cell interactions
Immunotherapies against PD1 and CTLA-4 are effective for the treatment of multiple tumour types and lead to durable responses for some patients. However, these therapies – referred to as checkpoint blockade – are not effective for all cancer types and the duration of responses remains poorly understood. The pre-clinical development of new immunotherapies is difficult because of a lack of human models systems that effectively mimic the interaction of the tumour and immune system.
We are developing new in vitro co-cultures models of patient-derived organoids together with immune cells to study the cellular factors that mediate immune cell mediated tumour cell killing.
The development of these models, and their use for the study of tumour-immune cell interactions, could lead to a deeper understanding of factors influencing patients responses to checkpoint blockade and new therapuetic hypotheses.
Core team

Syd Barthorpe
Senior Scientific Manager

Alexandra Beck
Project Manager

Mr Shriram G Bhosle
Principal Software Developer

Dr Olli Dufva
Postdoctoral Fellow

Dr Emma Duncan
Technical Specialist

Maria Garcia-Casado
Research Assistant

Dr Frederik Gibson
Technical Specialist

Prahalad Giridhar
PhD Student

Dr Carmen Herranz-Ors
Postdoctoral Fellow

Dr Liliya Kopanitsa
Advanced Research Assistant

Dr Howard Lightfoot
Principal Bioinformatician

Dr Katrina McCarten
Postdoctoral Fellow

Dr Clare Pacini
Principal Bioinformatician

Dr Saroor Patel
Senior Staff Scientist

Gabriele Picco
Postdoctoral Fellow

Mamta Sharma
Senior Technical Specialist

Rebecca Shepherd
Senior Software Developer

Samantha Walker
Research Assistant
Previous core team members

Mrs Angham Al Saedi
Advanced Research Assistant

Rizwan Ansari
Advanced Research Assistant

Grace Battarbee
Research Assistant

Dr Fiona Behan
Postdoctoral Fellow

Graham Bignell
Senior Staff Scientist

Sophie Brocklesby
Research Assistant

Jessica Cantwell
Research Assistant

Elisabeth D. Chen
PhD Student

Dr Matthew Coelho
Cancer Research UK Career Development Fellow

Dr Thomas Cokelaer
Senior Bioinformatician

Hristina Grigorova Dimitrova
Research Assistant

Dr Cansu Dinçer
PhD Student

Dr Lisa Dwane
Postdoctoral Fellow

Luke Farrow
Research Assistant

Hayley Francies
Senior Staff Scientist

Mr James Gilbert
Senior Software Developer

Emma Goffin
Former Advanced Research Assistant at the Sanger Institute

Emanuel Gonçalves
Postdoctoral Researcher

Caitlin Hall
Research Assistant

Dr Patricia Jaaks
Staff Scientist

Maciej Jablonski
Research Assistant

Megan Jukes
Research Assistant

Genevieve Leyden
Research Assistant

Ermira Lleshi
Research Assistant

Mr Iman Mali
Research Assistant

Emily Mallett
Research Assistant

Mr Kieron May
Research Assistant

Ms Anne Maria McLaren-Douglas
Senior Research Assistant

Tatiana Mironenko
Senior Research Assistant

James Morris
Research Assistant

Dr Romina Oliveira Silva
Staff Scientist

Riccardo Panero
Postdoctoral Fellow

Rachel Pooley
Research Assistant

Stacey Price-Weight
Head of Technical Development - DNAP

Mrs Laura Richardson
Advanced Research Assistant

Mr Gabriel Robert-Tissot
Research Assistant

Dr Marie Shamseddin
Postdoctoral Fellow

Mr Massimiliano Jacob Spensley
Research Assistant

Ms Fran Thomas
Research Assistant

Charlotte Tolley
Advanced Research Assistant

Dr Sara Valentini
Genomic Surveillance Business Analyst and Continuous Improvement Lead

Sara Vieira
Advanced Research Assistant

Mr Alex Watterson
Advanced Research Assistant

Wendy Yang
Senior Software Developer
Associated research
Related groups
Programmes and Facilities
Partners
We work with the following groups
External
Wellcome Trust
External
Open Targets
External
Cancer Research UK
External
Stand Up To Cancer
External
British Lung Foundation