Gene Editing
Cellular Operations
About us
- Through the adoption and implementation of modern genome editing techniques, we tailor our technical experience to help answer biological questions.
- We optimise, develop and democratise the delivery of genome editing tools and platforms for the Institute’s research programmes.
- For our collaborating partners we provide an agile, project focused, cost effective and efficient service as well as develop and provide biological resources, technical support and training for research groups and their staff.
Background
Gene editing (or genetic engineering) is a group of technologies that provide the ability to insert, delete, modify or replace the DNA in a living organism. In research, site specific gene modification is an established method to interrogate the function of genes (or gene products) in cells and organisms in both health and disease.
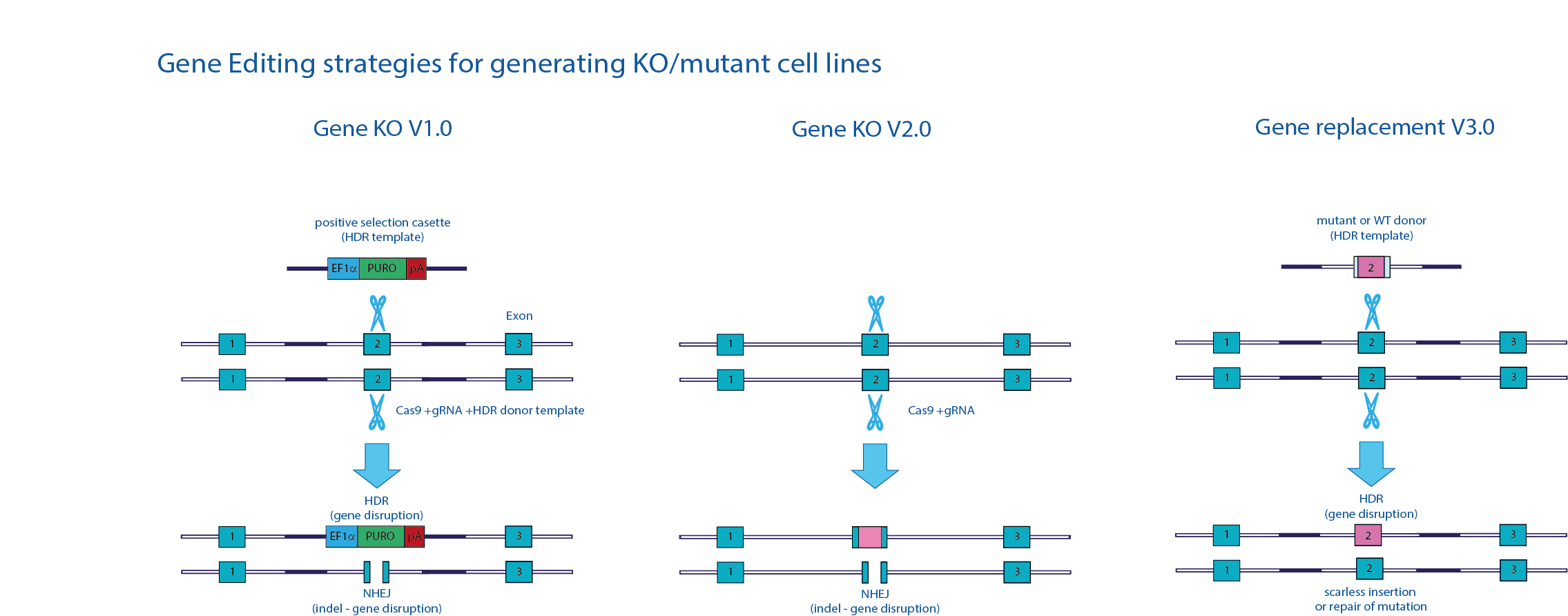
Revolutionary developments in genome editing techniques and methodologies have vastly improved the ability of researchers to perform site specific genetic engineering. In particular, the use of DNA nucleases such as ZINC fingers (ZFNs), transcription activator-like effectors (TALENs), and more recently the bipartite CRISPR-Cas system, have been key proponents to advancements in the field. These engineered nucleases, often referred to as ‘molecular scissors’, combine site specific genomic recognition with their ability to ‘cut’ DNA. Once DNA is cleaved the natural repair pathways of a cell attempt to repair the damaged DNA sequence. Repair via non-homologous end joining (NHEJ) can result in unsolicited insertions or deletions of nucleotides at the site of DNA cleavage and may cause disruption of normal gene function (i.e. loss of function). When paired with a repair or donor template (a fragment of DNA with known, predetermined sequence) precise insertion of DNA, via homology directed repair (HDR) can be achieved, such as the introduction of a specific mutation.
Resources
The ever expanding gene editing ‘tool-box’ enables researchers to permanently edit genes as well as perturb or tweak gene expression through the use of dCas9-fusion proteins.
The Gene Editing team routinely acquires, develops and tests novel CRIPSR-Cas and other gene editing reagents, resources which contribute to the Institute’s wider research goals.
Gene Editing Platforms
Building on the work of previous Sanger alumnus, Prof. Bill Skarnes and in concert with the Cellular Operations Research and Development team, lead by Andrew Bassett, we have developed a number of pipelines that establish loss of function (gene ablation) or precise genetic mutant cell lines.
These are used to model the effects of normal gene disruption in health and disease, such as those observed in developmental disorders (https://www.sanger.ac.uk/collaboration/deciphering-developmental-disorders-ddd). These work-flows have been optimised in the generation of some 1,400 mutant induced pluripotent stem cells (iPSCs). More recently, work has begun on expanding the use of these platforms to edit the genomes of primary human cell types, including cells of the haematopoietic system.
Allied to our efforts using CRISPR-Cas we also use well developed platforms to generate large numbers of traditional gene targeting vectors for organisms not amenable to gene targeting using CRISPR or TALENS, such as the malaria parasite P. berghei.
Large Scale Genetic Screening
CRISPR/Cas genome editing tools are extremely amenable to multiplexing and as such are the ‘go to’ tool for performing genome wide genetic perturbation screens. Via pooled or arrayed screening research teams are identifying candidate genes or genetic interactions responsible for a particular phenotype or disease susceptibility. This ability to inform genotype – phenotype interactions in a high throughput manner is empowering the large scale identification of biological networks and the next level of drug discovery. These are also powerful tools to examine the direct effects of a gene, gene variant or gene perturbation on a biological outcome and when combined with single cell –omics, a genotype – phenotype relationship can be determined at the level of a single cell.
The Gene Editing team has developed platforms for generating both arrayed and pooled libraries. In close collaboration with the Cellular Operations Research and Development team, the wider Institute Faculty and external collaborators we are involved in the evolution of these technologies and aim to further develop their design, generation and delivery.
We also have expertise in the use of transposons, genetic elements which can be mobilised in a cell or organism by enzymes known as transposases. Once mobilised, these elements insert randomly into an organisms genome and can activate or disrupt transcription of neighbouring genes (i.e. random insertional mutagenesis).
Our people
Core team

Mrs Michaela Bruntraeger
Senior Research Assistant

Mr David Fraser
Advanced Research Assistant

Miss Vimbai Sharon Madzorera
Advanced Research Assistant

Dr Sophie Robinson
Advanced Research Assistant
Previous core team members

Dr Meg Byrne
Senior Research Assistant

Dr Oliver M Dovey
Head of Gene Editing, Cellular Operations

Ben Newman
Advanced Research Assistant

Tabea Nimz
Advanced Research Assistant

Dr Mark Thomas
Principal Bioinformatician

Eloise Wells
Advanced Research Assistant