Bayraktar Group
Cellular Genomics
We seek to explore the vast cellular diversity in the human body.
To understand human cellular diversity, we aim to
- create single-cell level 3D maps of human organs using large-scale spatial transcriptomics and
- uncover the specialized functions of brain cell types using large-scale in vitro screens and mouse models.
We focus on studying neural cell type diversity in the human cerebral cortex
Without the cerebral cortex, we would not have complex thought and behavior. The cortex also plays major roles in many neurodevelopmental and psychiatric disorders including autism spectrum disorders (ASDs) and schizophrenia. To precisely understand cortical function and determine how it goes awry in disease, we need to classify more than 16 billion neurons and 60 billion glial cells across this complex brain structure.
Research directions
Our team will work on three major research directions:
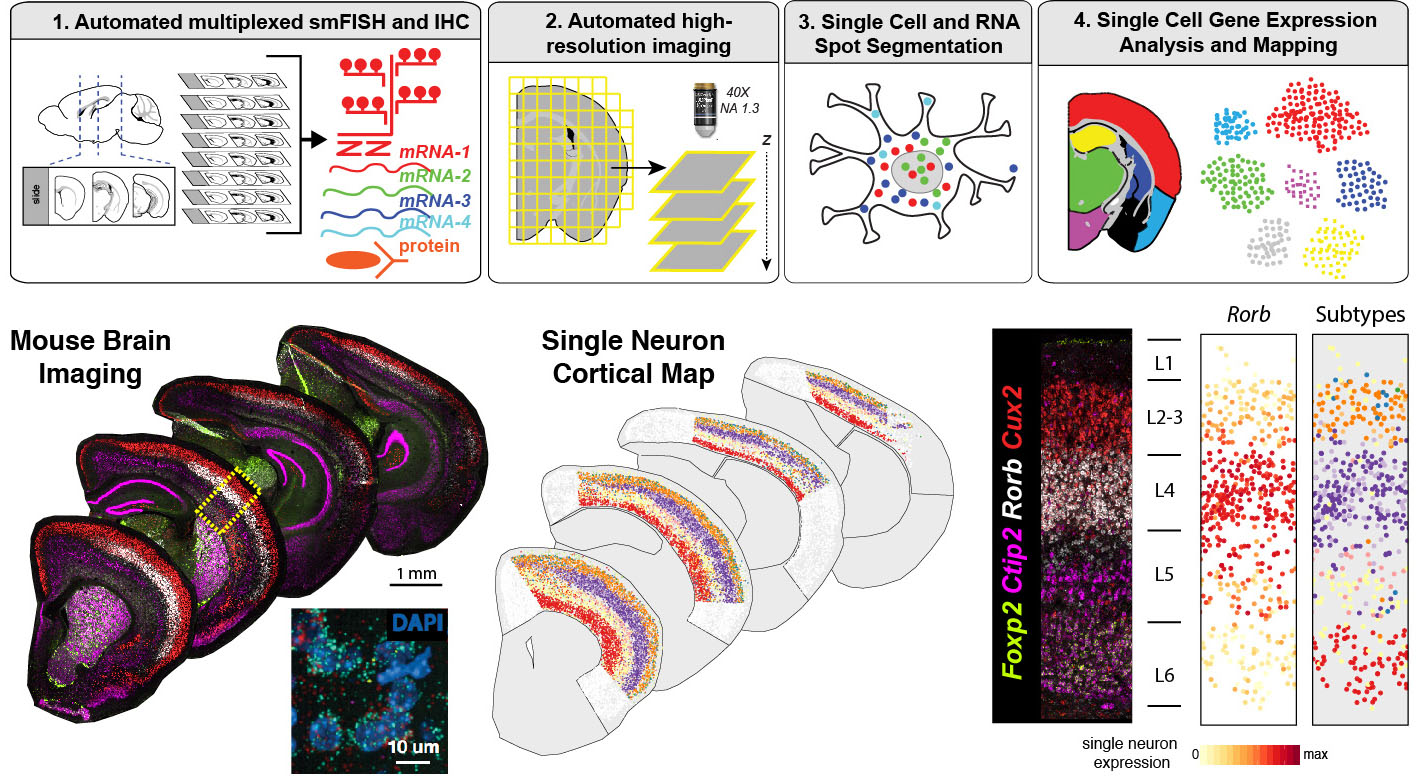
- Large-scale spatial transcriptomics for mapping human tissues: We will establish automated histology and imaging pipelines to map human tissues at single cell resolution at scale. We will use and develop highly-multiplexed single molecule fluorescent in situ hybridization (smFISH) methods to identify molecular cell types. We will extensively collaborate with Human Cell Atlas (HCA) and other teams at Sanger on diverse tissue applications and to develop automated image data analysis pipelines.
- Cortical cell type diversity in health and disease: We will use single cell sequencing and large-scale spatial transcriptomics to map neuronal and glial subtypes in the developing and adult human cerebral cortex. We will further utilize spatial transcriptomics to identify cellular pathways involved in neurodevelopmental disorders such as ASD.
- Large-scale screens to discover human glial function: Glia represent the majority of cells in the human cerebral cortex, but we know little about their biology. We will perform large-scale protein interaction screens and imaging-based cellular assays to discover glial molecular pathways that regulate neuronal development. We will also study glia-neuron interactions in vivo using mouse models. In our initial studies, we will focus on the role of human astrocytes in synapse development.
Core team

Sabine Eckert
Staff Scientist

Dr Loren Gibson
Business Analyst and Process Improvement Lead

Oliver Gould
PhD Student

Jing Eugene Kwa
PhD Student

Dr Kwasi Amoako Kwakwa
Visiting Scientist

Dr Tong LI
Senior Software Developer

Dr Jimmy Tsz Hang Lee
Senior Data Scientist

Dr Fani Memi
Senior Staff Scientist

Mathieu Perez
MD-PhD Candidate

Tarryn Porter
Senior Scientific Manager

Mr Koen Rademaker
Bioinformatician

Dr Kenny Roberts
Senior Staff Scientist

Dr Qianqian Zhang
Postdoctoral Fellow
Previous core team members

Dr Gavin Bowen-Metcalf
Advanced RA

Dr Yeliz Demirci
Postdoctoral Fellow

Erdem Ercan
Summer Student Placement

Dr Vitalii Kleshchevnikov
Bioinformatician

Dr Jun Sung Park
Visiting Scientist

Dr Benjamin (Ben) John Woodhams
Postdoctoral Fellow in High Throughput Spatial Genomics
