Wright Group
Cell Surface Signalling Laboratory
Archive Page
This page is maintained as a historical record and is no longer being updated.
Gavin Wright moved to the University of York in 2021. To find out more about his team’s ongoing research, please visit: https://www.york.ac.uk/biology/our-staff/gavin-wright/ and https://pure.york.ac.uk/portal/en/researchers/gavin-james-wright(7c9756f5-9f6c-42ef-877b-bffc627cdafd).html
The focus of our research is the molecules which are displayed on the surface of cells, and in particular, the extracellular binding events that they mediate. Extracellular interactions are excellent therapeutic targets because they are directly accessible to systematically delivered drugs such as monoclonal antibodies. For infectious diseases, proteins displayed on the surface of pathogens are good vaccine candidates because vaccine-elicited antibodies can destroy or disable the pathogen.
We use a mammalian expression system to produce large panels of soluble recombinant proteins to increase the chances that the proteins will be correctly folded and active; the proteins are then used for protein interaction and preclinical vaccine screening. We have developed infrastructures that allow us to express and purify many hundreds of recombinant proteins expressed in this system rapidly and conveniently.
New methods to identify low affinity extracellular receptor-ligand interactions

We have invented and continue to develop methods for large scale extracellular interaction discovery such as our AVEXIS technique. Currently, we are developing genome-scale receptor screening systems based on both creating transfected cellular microarrays, and collaborate with other teams at the Institute to use genetic approaches based on the CRISPR-Cas9 method. One unifying aspect of these methods is that we are expecting extracellular interactions to be very weak, and so we purposefully increase binding avidity when performing our receptor interaction screens experiments by oligomerising the protein probes.
Systematic preclinical vaccine screens for neglected tropical diseases
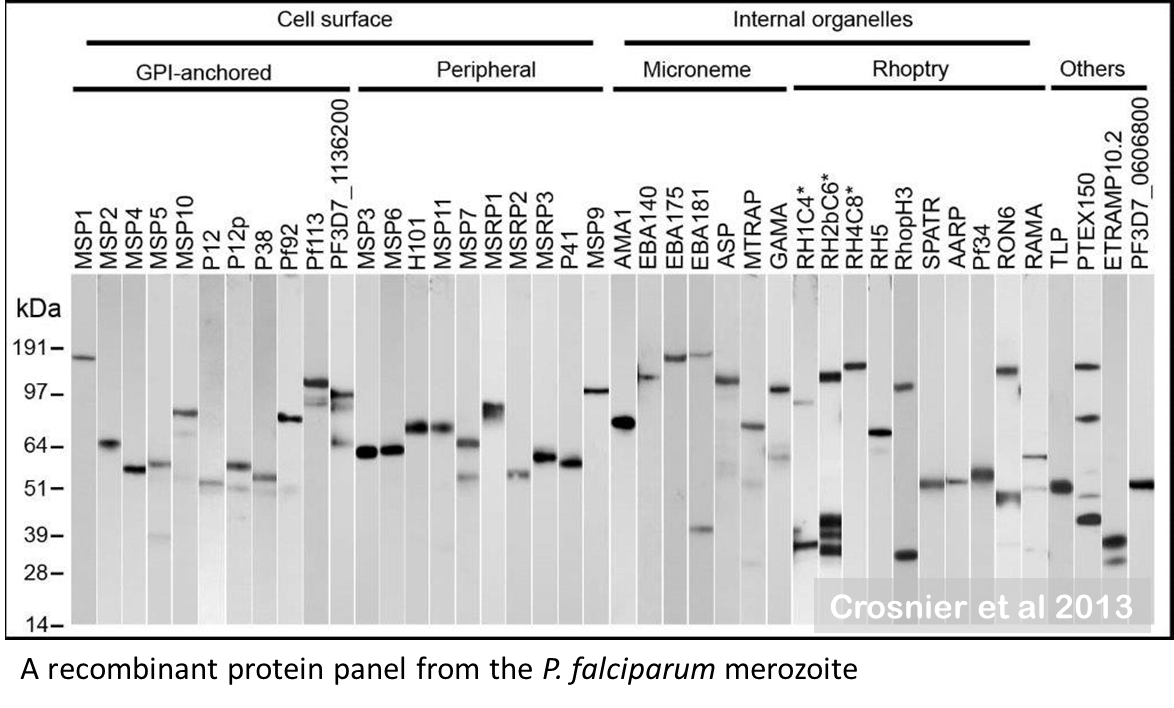
We use our mammalian expression technologies to create panels of recombinant pathogen proteins that are then used to identify new subunit vaccine targets for infectious diseases. We have compiled a large panel of proteins representing the cell surface receptor repertoire and secreted proteins from the blood stage of the malaria parasite.
These proteins can be comparatively tested to identify the most promising vaccine candidates, and to understand how the parasite interacts with the human host and cause disease. As well as malaria, we are working on other parasitic diseases including schistosomiasis, leishmaniasis and trypanosomiasis – infectious diseases that affect some of the most disadvantaged people in world.
Videos of some of our research
Previous core team members

Dr Delphine Autheman
Postdoctoral Fellow

Enrica Bianchi
Staff Scientist

Shan Chong
Visiting Worker PHD

Dr Cecile Crosnier
Senior Staff Scientist

Kirsten Dundas
Former PhD Student at the Sanger Institute

Dr Francis Galaway
Postdoctoral Fellow

Dr Julia Knoeckel
Postdoctoral Fellow

Dr Michelle McRae
Senior Research Assistant/Laboratory Manager

Dr Han Ong
Postdoctoral Fellow

Dr Catherine Onley
Daphne Jackson Fellow

Sumana Sharma
PhD Student

Dr Laura Wood
Postdoctoral Fellow
Related groups
Programmes and Facilities
Partners
We work with the following groups

