WGE - CRISPR design tool
WTSI Genome Editing (WGE) is a website and database that provides tools for designing genome editing of human and mouse genomes using CRISPR/Cas9
About
General Introduction
WGE is a highly interactive, web-based visual tool that employs an embedded genome browser and database to assist scientists in designing genome editing strategies using the CRISPR/Cas9 system.
Genome editing is the science of altering specific portions of DNA so that it behaves in a subtly different way within a cell. For example, we can use genome editing to prevent specific genes from being translated and subsequently transcribed into functioning proteins. In this way we can examine the role genes play in the life of the cell, potentially shedding valuable light on the causes of disease.
Proteins are the workhorse molecues of the cell, they provide physical structure to cells as well as carrying out work by ensuring chemical reactions occur in the right order, using the correct amount of energy and the appropriate ancillary molecules. Even a single unit of a protein, known as an amino acid residue, that is wrong can lead to debilitating disease. For example, sickle cell anaemia is due to a single amino acid change that makes haemoglobin too sticky.
DNA is the molecule that carries the genetic code that specifies the sequence of amino acid resides in the protein products of the cell. It also carries a lot of information in non amino acid encoding regions that guides the way genes are expressed. Any of these regions could be the targets for genome editing campaigns.
Detailed Description
The rapid development of CRISPR-Cas9 mediated genome editing techniques has given rise to a number of online and stand-alone tools to find and score CRISPR sites for whole genomes. The Wellcome Trust Sanger Institute Genome Editing database (WGE), uses novel methods to compute, visualize and select optimal CRISPR sites in a genome browser environment. The WGE database currently stores single and paired CRISPR sites and pre-calculated off-target information for CRISPRs located in the mouse and human exomes. Scoring and display of off-target sites is simple, and intuitive, and filters can be applied to identify high-quality CRISPR sites rapidly. WGE also provides a tool for the design and display of gene targeting vectors in the same genome browser, along with gene models, protein translation and variation tracks. WGE is open, extensible and can be set up to compute and present CRISPR sites for any genome. [Hodgkins et al. 2015]
The WGE website presents two tools for assisting genome editing:
- a CRISPR finder
- a Gibson assembly PCR oligo designer
The CRISPR-finding part of the WGE website requires the user to select a species—currently human or mouse—and the marker symbol of the gene to inspect. The target Ensembl exon is then selected from a list. All possible CRISPR sites and paired sites are shown on a scrollable Genoverse genome browser view. This incorporates the current gene models from Ensembl, protein translation, available variation and any user-generated targeting vector designs. ‘Paired’ CRISPR sites for applications using Cas9 nickases are shown in WGE when CRISPR sites on opposite strands have a separation of up to 30 bp, or an overlap of up to 10 bp. [Shen et al. 2014]
Scoring Off-targets
Our scoring system reports the number of similar sequences in the genome with up to four mismatches (excluding the PAM region). A string such as ‘0:1, 1:0, 2:0, 3:4, 4:56’ indicates that there is 1 genomic site with 0 mismatches (the CRISPR site itself), no off-target sites with 1 or 2 mismatches, and an increasing number of potential off-target sites with 3 and 4 mismatches. In this way, users can immediately grasp the off-target potential for each CRISPR site.
Users can also filter CRISPR sites based on their stored off-target characteristics. Using this interface, hundreds of possible CRISPR sites can be narrowed down and evaluated to select the optimal site(s) for an editing task.
Gibson Assembly Designer
WGE can be used to design PCR primers for the assembly of gene targeting vectors by Gibson assembly or other similar PCR-based methods. This involves first choosing a target exon, and then adjusting design parameters via a web interface to allow the primer calculations to be run. The resulting targeting vector designs can be bookmarked, edited, and are displayed alongside CRISPR sites in the genome browser.
WGE contains comprehensive help and an application programming interface that enables users to build their own CRISPR indexes for specific projects.
See the publications below for further details.
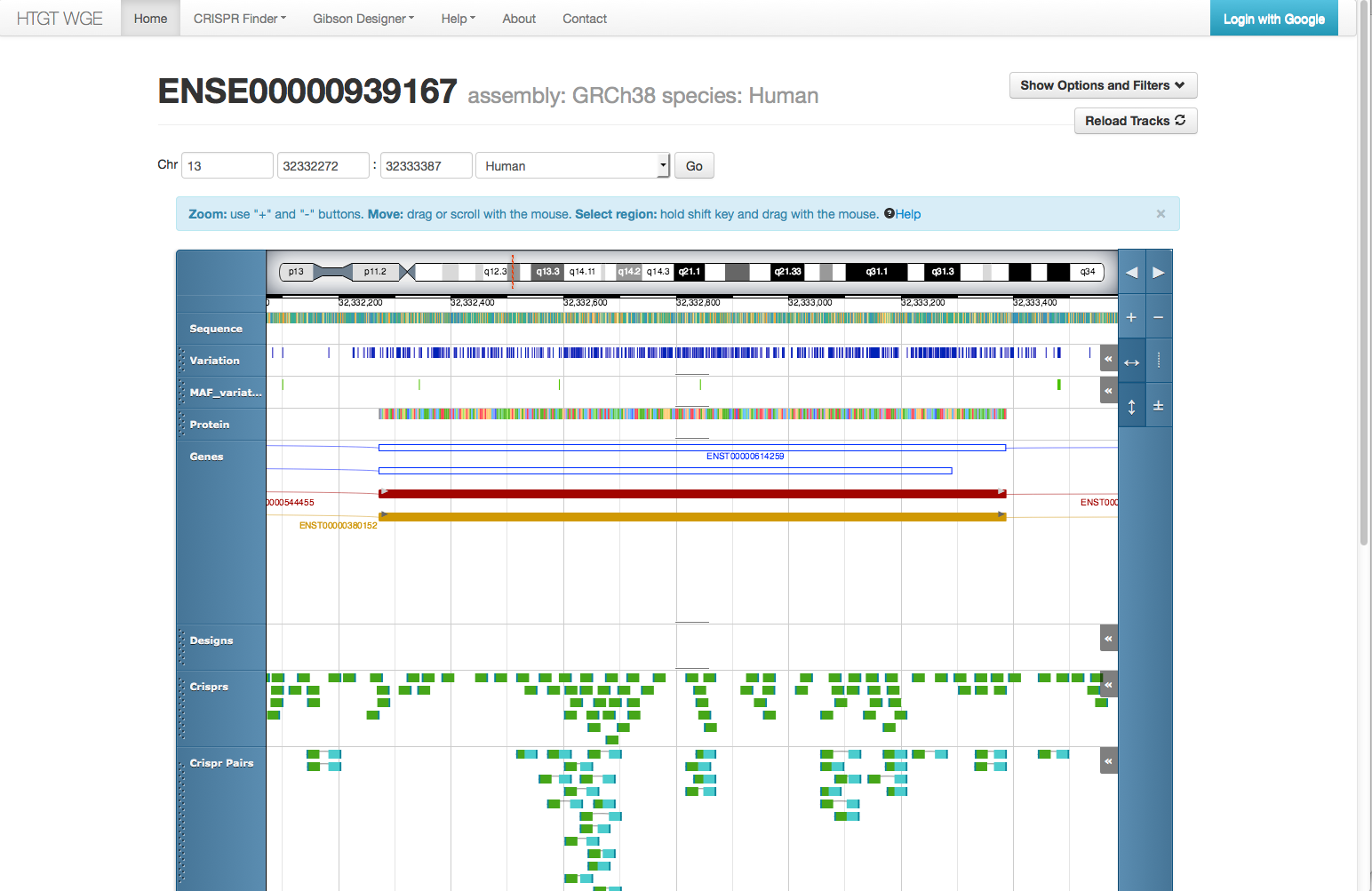
Working on selection of CRISPRs for a genome editing experiment in the WGE Genome Browser
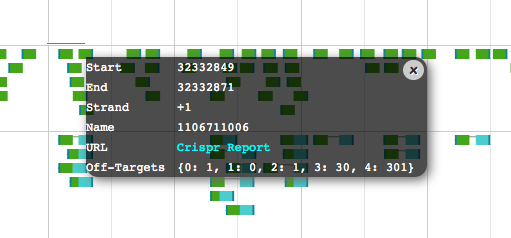
CRISPR context menu. The 'Crispr Report' link reveals detailed information on the selected CRISPR
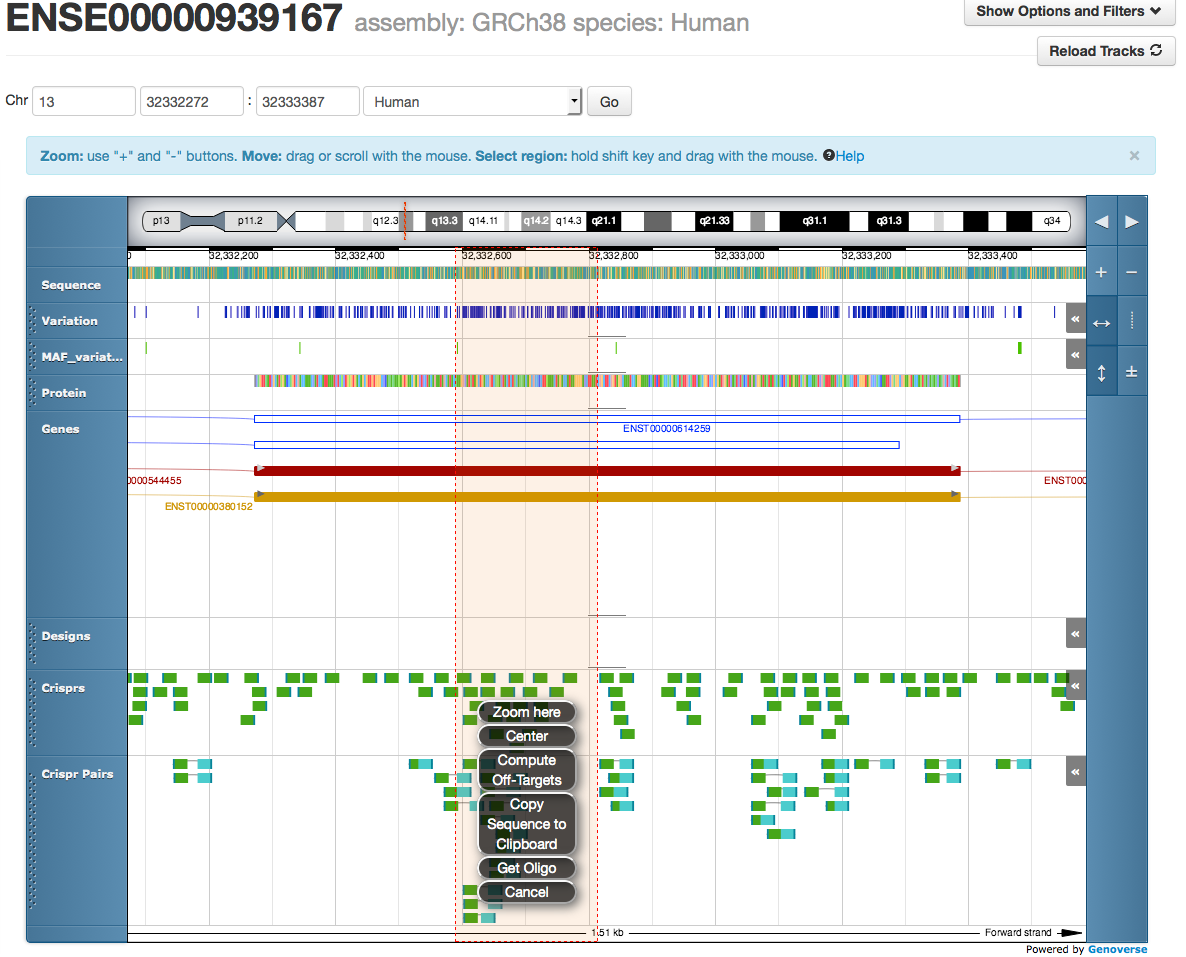
By shift-click-dragging, a range including CRISPRs and genomic sequence can be selected and acted upon.
Downloads
WGE should be accessed through the WGE website. The source code is available at Github.
The WGE API should be used to search the CRISPR database for mouse or human CRISPRs.
The CRISPR-Analyser code is available if you want to create your own database of CRISPR sites for any genome.
Further information
You are strongly advised to read the online help in WGE as this contains a detailed tutorial that will enable you to get the best from your CRISPR design efforts. If you need help after reading this document please send your request to our help queue.
Recent improvements include the ability to copy sequence from the web browser to a clipboard for custom CRISPR and PCR design strategies. This is available when you shift-click-drag to select a region in the Genoverse browser. Once you release the click, the region is selected and a menu appears including the new Copy Sequence to Clipboard function. See Screenshots to the right of this text.
WGE and its supporting codebase is Copyright 2015 Genome Research Limited and is open source software available under the GNU AGPL v3 licence. The CRISPR-Analyser code base that generates our CRISPR on and off-target database is available under the MIT licence.
To cite this work please use our 2015 Bioinformatics paper. See reference below.
Contact
If you need help or have any queries, please contact us using the details below.
All enquiries about WGE should be sent to our ticketing system at wge@sanger.ac.uk
The maintainer of this page is David Parry-Smith
This tool is maintained and supported by the Stem Cell Informatics team.
Sanger Institute Contributors

Vivek Iyer
Human Genetics Informatics Team Lead
Previous contributors

Tiago Grego
Software Developer

Dr David Parry-Smith
Head of Cellular Informatics

Dr Bill Skarnes
Former Senior Group Leader
External Contributors

Alex Hodgkins
Contributed to WGE while working at the Sanger Institute.

Saj Perera
Contributed to WGE while working at the Sanger Institute.