High Throughput Spatial Genomics
Sanger-EBI Initative for Large-Scale Spatial Genomics
Mission Statement
Recent technological advances in spatially resolved RNA-sequencing and high-multiplexed RNA imaging have pioneered the spatial transcriptomic analysis of tissues. The promises of these exciting spatial transcriptomics technologies – 3D cellular atlases of entire tissues/organisms and spatial biology of hundreds of patient samples – can only be realized with large-scale experimental and computational platforms.
The High-Throughput Spatial Genomics (HTSG) initiative utilises cutting-edge technologies to generate spatial omics data at scale. We combine single cell and spatial RNA-sequencing, high-throughput microscopy and quantitative image data analysis to enable large-scale spatial genomic studies. Our activities strongly tie into the Human Cell Atlas as well as genomic technology development and cancer biology.
Spatial Technologies
The Initiative employs diverse sequencing and imaging technologies for spatial transcriptomic data generation.
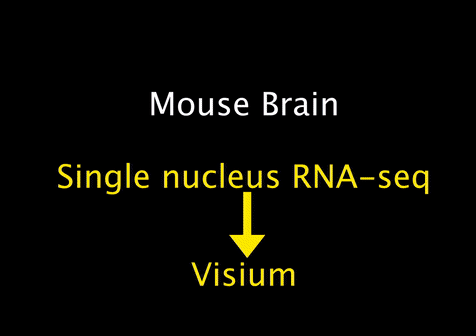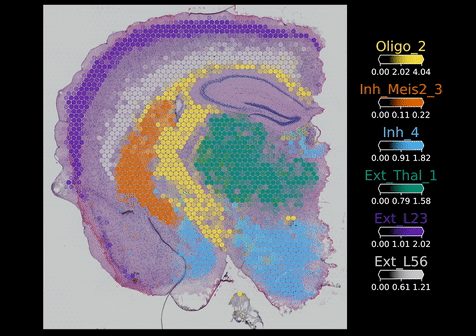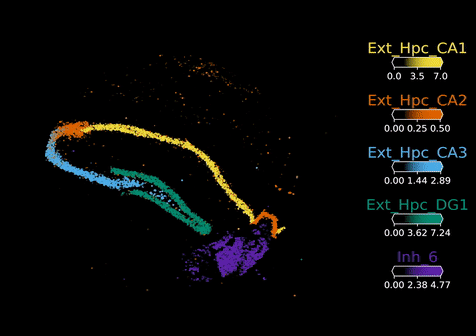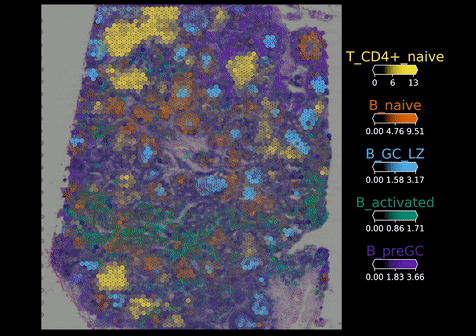
Visium Spatial Transcriptomics:
Visium technology (10X Genomics) enables spatial RNA-sequencing (RNA-seq) by positionally capturing RNAs from thin tissue sections onto an oligonucleotide array. The Visium arrays provide a spatial resolution of 55 microns – capturing multiple cells at each tissue position. To computationally extend the resolution of the Visium assay and other spatial RNA-seq methods to map individual cell types, we developed the cell2location model. Cell2location integrates single cell and spatial transcriptomic data to comprehensively map cell types across complex tissues.
We utilise both Visium for fresh frozen samples, and the probe hybridisation-sequencing hybrid Visium CytAssist for FFPE tissues.
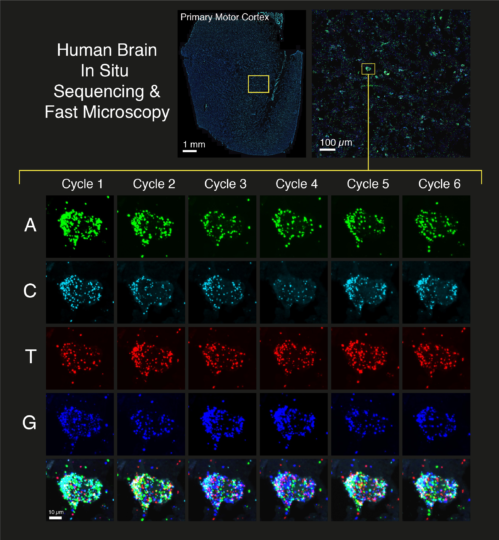
In Situ Sequencing and Xenium:
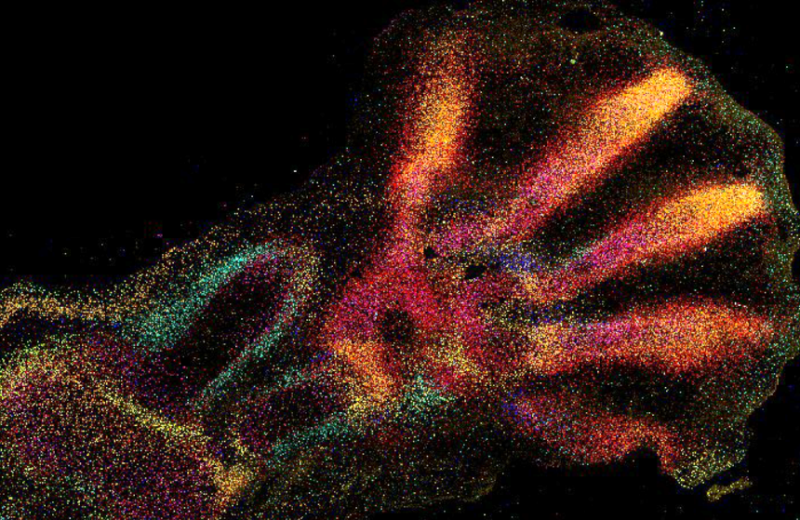
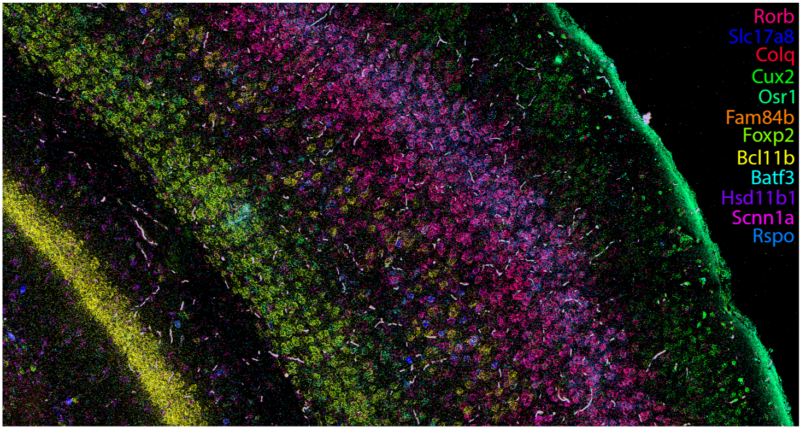
We use both manual in situ sequencing (CARTANA) and automated 10x Genomics Xenium to quantify gene expression at single-cell resolution in tissues. In situ sequencing (ISS) provides a cyclic and combinatorial barcoded FISH approach to label up to 500 different transcripts in a multiplexed manner. We have coupled ISS with fast tissue imaging and bespoke image processing pipelines to generate a high-throughput pipeline for various human tissues.
Hiplex RNAscope:
This protocol provides highly-sensitive measurements of small gene panels in tissues. Hiplex RNAscope provides a cyclic and signal amplified FISH approach to label up to 12 transcripts in a multiplexed manner.
Instrumentation
Perkin Elmer Opera Phenix HCS
We have adapted this High Content Screening system for automated imaging of tissue sections labeled with ISS or RNAscope smFISH. To provide high-throughput imaging at high spatial resolution, this system was assembled with a Yokogawa spinning disk and 4 cameras as well as automated water immersion objectives and software autofocus.

CAIRN “Nemo” Spinning Disk
The most recent addition to the facility, “Nemo” is a custom dual-camera spinning disk confocal microscope assembled around a CrestOptics X-light V3 by Cairn Research. The completely open hardware and software the microscope runs on gives the facility more flexibility to implement automated cyclic imaging protocols for ISS and other high-multiplexed RNA imaging methods.
CAIRN “Ahab” (coming soon)
The next planned instrument for the facility will be a larger, four-camera version of “Nemo” also to be assembled and installed by Cairn Research
Image Data Infrastructure
Cyclic Microscopy Image Analysis Pipelines
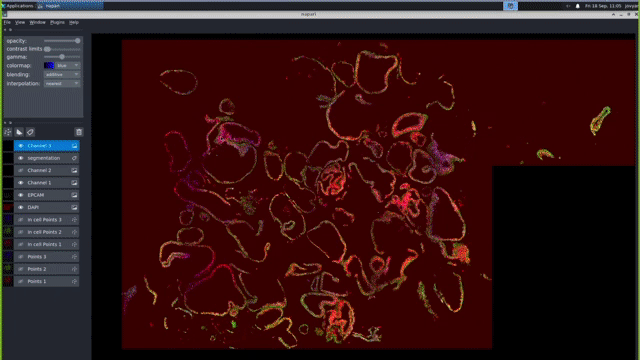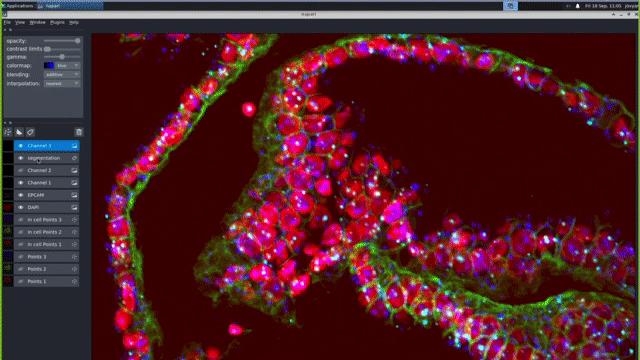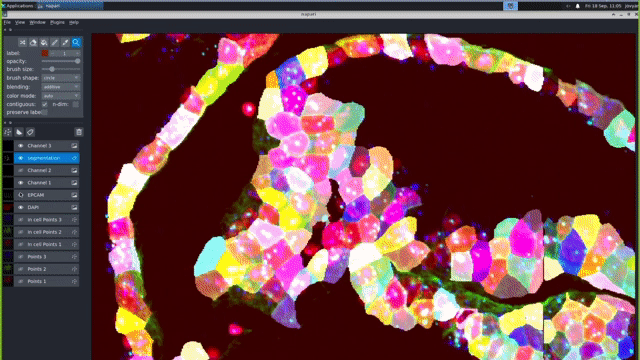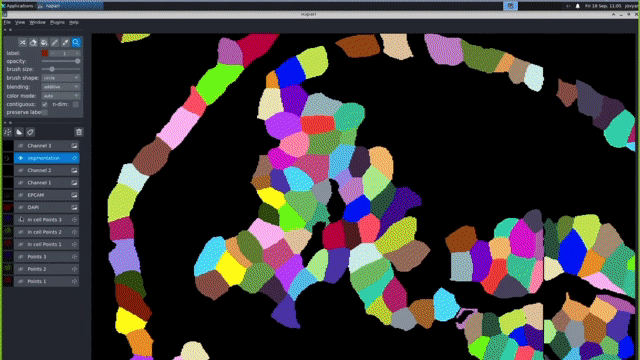
Our computational pipelines and IT platforms enable processing ISS & RNAScope image datasets at scale. Our bespoke workflows enable registration of large cyclic images to sub pixel accuracy, robust barcode decoding and cellular segmentation across a variety of different organs and organoids. We use python based tools cell segmentation (e.g. cellpose) and visualisation (napari). To enable large-scale application, we deploy nextflow/jupyter based pipelines on high performance GPUs. These IT activities are supported by the Cellgen IT and Scientific IT teams at Sanger.
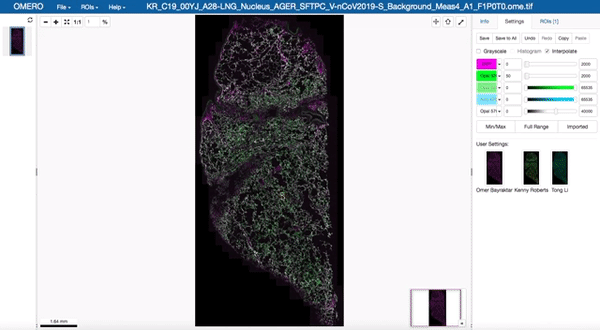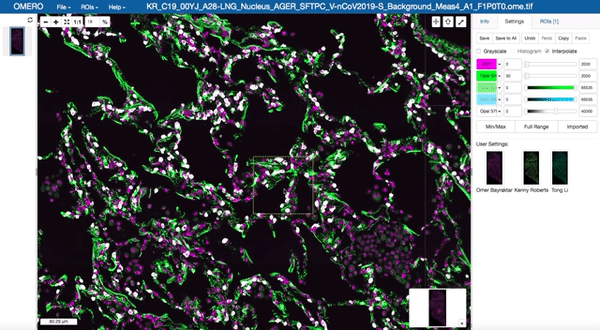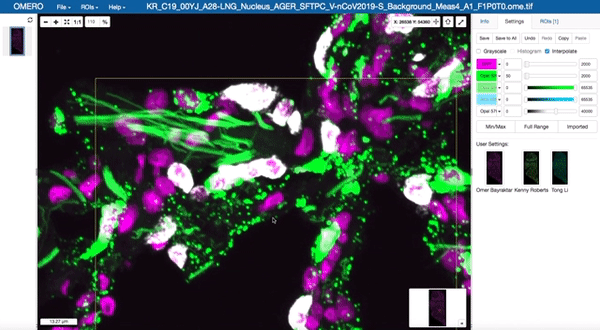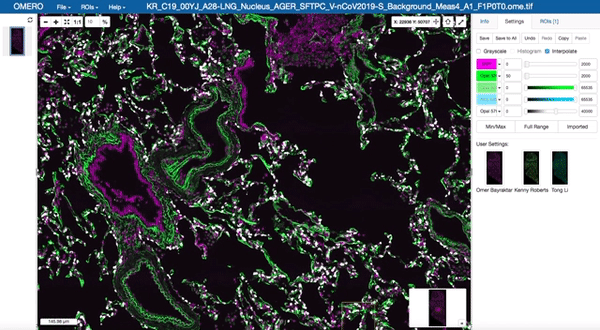
Image data management and visualisation via OMERO
All image data produced by the facility is stored in our OMERO databases maintained by the Cellgen IT team. Our Image Data Resource (IDR) instances allow us to visualise and browse large tissue image datasets in the “Google Maps” format and also enable us to share image datasets with external collaborators. These databases are maintained on petabyte-scale high performance storage systems.
BioImage Archive
We collaborate with the new BioImage Archive to disseminate our large-scale spatial transcriptomic image datasets to the researcher community.
Core team

Dr Kwasi Amoako Kwakwa
Visiting Scientist

Dr Tong LI
Senior Software Developer

Dr Fani Memi
Senior Staff Scientist

Dr Kenny Roberts
Senior Staff Scientist
Previous core team members

Dr Jun Sung Park
Visiting Scientist

Aleksandra (Ola) Tarkowska
Solutions Architect

Dr Benjamin (Ben) John Woodhams
Postdoctoral Fellow in High Throughput Spatial Genomics
