Malaria Parasite Genomic Surveillance
Genomic Surveillance Unit
Archive Page
This page is maintained as a historical record and is no longer being updated.
About us
We were biologists, bioinformaticians, technicians, and data scientists working at the intersection of computation, biology, lab methods, and public health. With a strong emphasis on creating useful data for our public health partners, we sought to understand the malaria parasite and its evolution. We worked collaboratively with other teams in the Genomic Surveillance Unit and around the world to scale the extraction of useful, high-quality genomic data from malaria parasites.
Our work
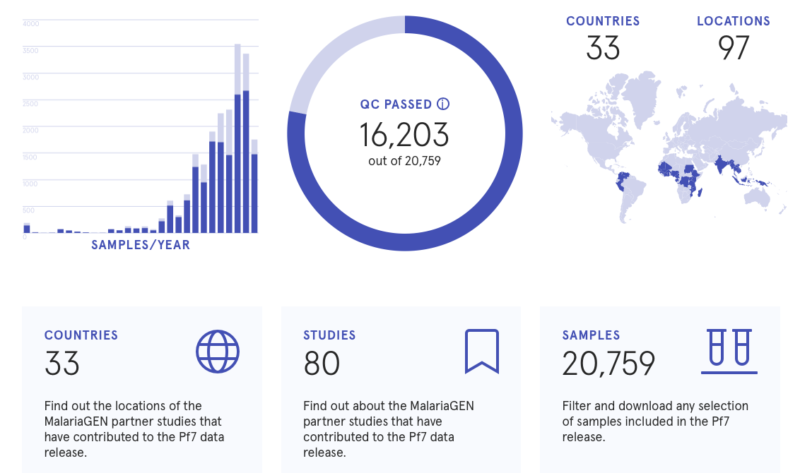
Building on nearly twenty years of experience integrating with the MalariaGEN network, we created data resources that track genetic changes in Plasmodium parasites, mostly of the two species responsible for most malaria deaths: P. falciparum and P. vivax. The largest of these resources is Pf7, a dataset containing more than 20,000 whole genome sequences of P. falciparum parasites from 33 countries. The P. vivax version of that dataset, Pv4, contains nearly 2,000 whole genome sequences.
We helped build up surveillance capacity in malaria-endemic areas through projects like the NIHR-funded Genomic Surveillance Hubs in West Africa Global Health Research Group and by supporting partners across the world in their genomic surveillance efforts.
Building on successes with amplicon sequencing for tracking antimalarial drug resistance, we built an Amplicon Toolkit with other teams in the Genomic Surveillance Unit. This suite of tools and services allowed partners to implement amplicon sequencing for genomic surveillance of malaria parasites. It was an end-to-end product that included metadata curation, training, sequence upload, bioinformatic analysis, data return, and genetic report cards.
Our team’s lab experience helped create new ways to make sequencing the parasites easier. For example, we looked at how to enhance Plasmodium vivax DNA within samples to be able to look at this parasite in more detail and how to extract DNA from malaria rapid diagnostic tests, essentially a waste product from malaria diagnosis. We also researched and developed also techniques to access complex parts of the parasite genome using long read sequencing technologies.
Our people
Previous group lead

Dr Cristina Ariani
Malaria Parasite Surveillance Lead
Core team

Dr William L. Hamilton
Clinical Lecturer in Medical Microbiology

Dr Eyyub Unlu
Data Scientist
Previous core team members

Dr Kim Judge
Research Software Engineer

Georgia Elizabeth Whitton
Senior Data Scientist