
AI-SEED-RCC: Using generative AI to spatially map evolution, environment, and drug responses to renal cell carcinoma
Kidney cancers behave very differently between patients. At the moment it very difficult to predict which tumours will develop aggressively and which will respond to therapy. This means some people miss out on receiving early treatment while others have interventions they don’t need.
The AI-SEED-RCC collaboration views cancer as more than just a problem of the faulty cells that become tumours (“the seed”). It also seeks to explore the environment around those faulty cells (“the soil”), such as immune cells and blood vessels, that determine whether the cancer grows quickly or slowly.
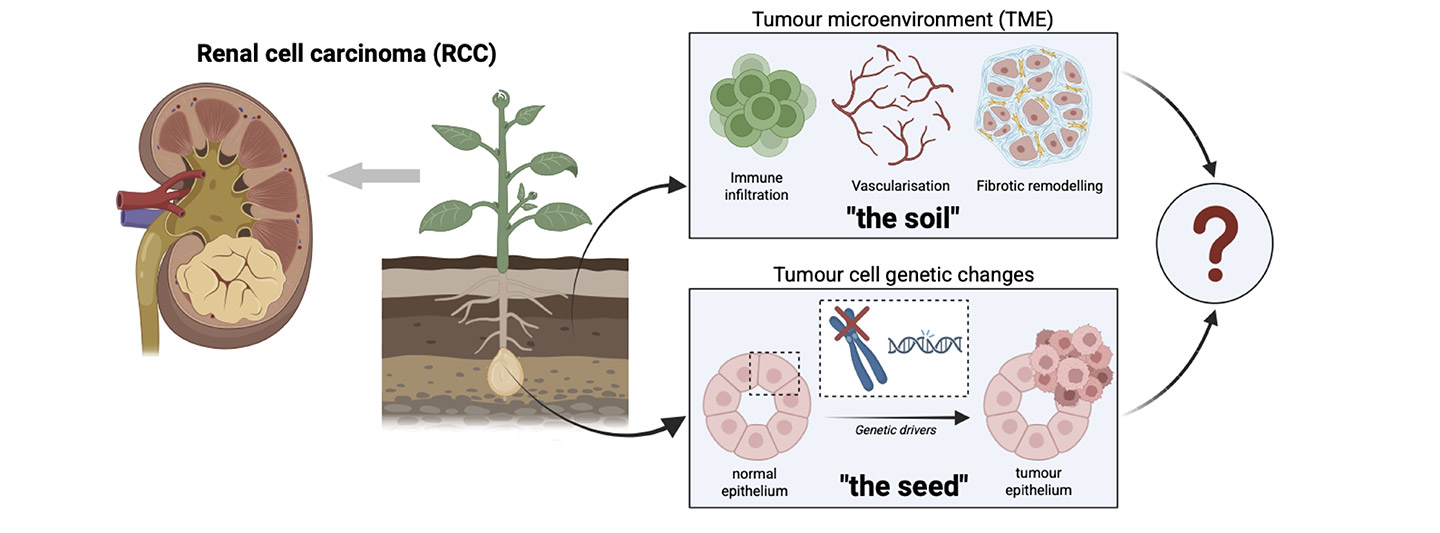
Exactly how “seed” and “soil” interact across patients remains a major unanswered question. These interactions are highly spatial, evolve over time, and cannot be easily perturbed in human tissue.
We combine spatial genomics and newly developed generative AI models to dissect how tumour “seeds” (genetic alterations) and “soil” (the tumour microenvironment) drive progression and therapy response in renal cell carcinoma (RCC).
Specifically, we develop AI models that learn the rules by which tissue context reprograms cell states, disentangle intrinsic tumour programs from microenvironment-induced effects, and predict how tumour ecosystems change under perturbation. Generative AI models such as MintFlow (Akbarnejad A. et al. Mapping and reprogramming human tissue microenvironments with MintFlow. bioRxiv (2025). doi:10.1101/2025.06.24.661094) provide the foundation for this approach.
The clinical issues AI-SEED-RCC seeks to address
By improving our understanding of kidney cancer in the three areas listed below, we hope to provide key information for developing better treatments and approaches:
- Lack of precision: It is difficult to know which early tumours will become dangerous and which will not, leading to overtreatment or missed opportunities to act early.
- Treatment resistance: Many patients stop responding to existing drugs, in part because tumour biology can adapt over time and in response to its surrounding tissue environment.
- Limited understanding of the tumour environment: It is unclear how cancer cells interact with the vasculature supplying these tumours and the immune cells which attempt to fight them, and which of these interactions are causally important.
Aims
To provide the knowledge required to improve kidney cancer care, the AI-SEED-RCC collaboration seeks to:
- Create tumour maps:
We use spatial genomics and AI to analyse kidney tumours from untreated patients to create detailed maps that track how cancer cells and their genetics change, and explore how their surroundings behave and change as the disease progresses. - Track responses to treatment:
Using samples from unique clinical trials, we study how tumours respond to therapy over time, to understand why some respond while others develop resistance. - Predict and test new treatments:
Applying generative AI models developed within AI-SEED-RCC, we perform in silico perturbations of tumour tissues, simulating how alterations to “the seed” or “the soil” influence kidney cancer behaviour.
These models are explicitly designed to predict the impact of removing, replacing, or reprogramming specific cell populations, enabling identification of key regulators and candidate drug targets that are difficult or impossible to perturb directly in patients or conventional laboratory systems.
The results inform the design and testing of newer and smarter anti-cancer drugs in cutting-edge experimental models, including advanced 3D cultures and organ-on-chip systems.
AI-SEED-RCC combines spatial genomics and bespoke generative AI model development to dissect how tumour “seeds” (genetic variations) and “soil” (tumour microenvironment) drive progression and therapy response in RCC. Insights from longitudinal patient cohorts guide in silico tissue perturbation, which is then linked back to experimental validation to identify and test novel therapeutic strategies.
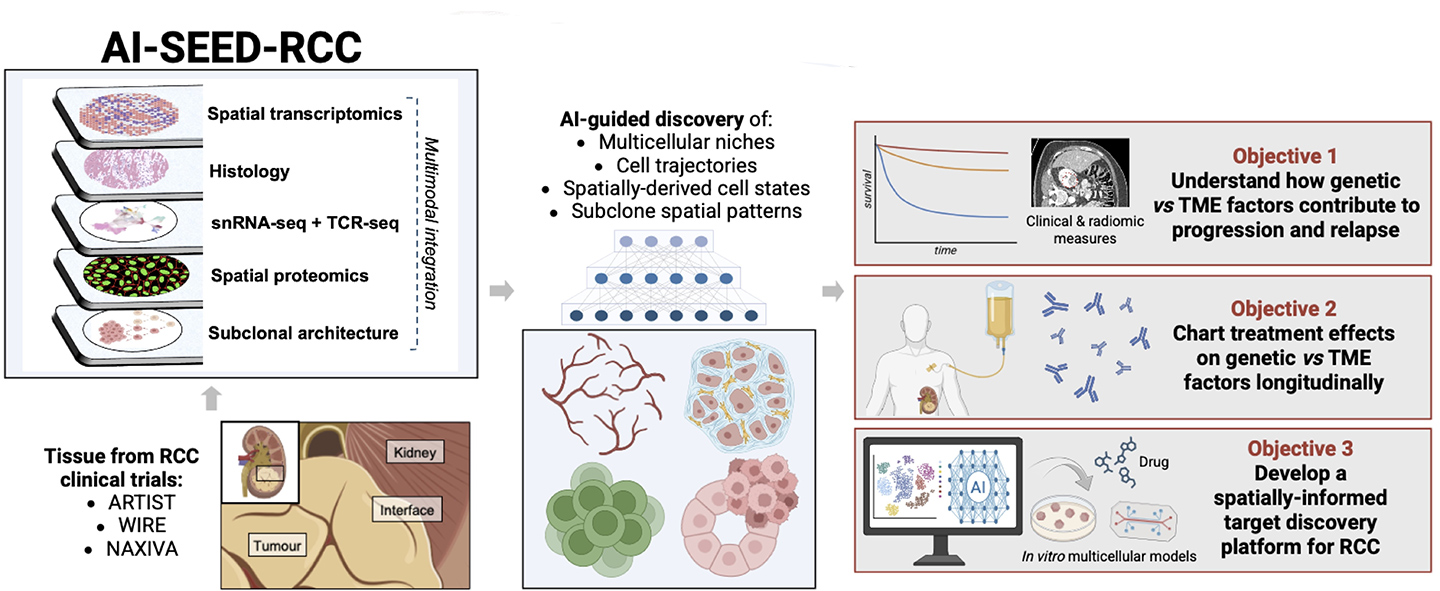
Why generative AI makes this possible
Traditional computational methods can describe tumour tissues, but they cannot predict how tissues will change when specific cells or signals are altered. This limitation arises because most approaches analyse observed data, rather than modelling the biological processes that generate tissue states.
Generative AI overcomes this barrier by explicitly modelling how tissue organisation and gene expression emerge from interactions between cells and their microenvironment. By developing AI models that learn these generative rules from patient-derived spatial data, AI-SEED-RCC can generate new, biologically plausible tissue states that were not directly observed.
This enables in silico tissue perturbation: virtually removing, replacing, or modifying specific cell populations within intact tumour ecosystems and predicting how the rest of the tissue responds. Such experiments are extremely difficult or impossible to perform in human tissue and cannot be explored systematically using standard in vitro or animal models.
Closing the loop: from virtual tissues to real biology
AI-SEED-RCC is designed to close the loop between computation, experimentation, and patient data:
- In silico (virtual tissues):
Generative AI models developed within the project are trained on spatial genomics and imaging data from patient tumours. These models are then used to perform in silico perturbations, identifying key regulators of tumour progression, immune suppression, and drug response. - In vitro (targeted experimental validation):
Predictions from in silico perturbation guide focused experimental testing in advanced laboratory systems, including 3D tissue models and organ-on-chip platforms. Instead of attempting to recreate full tumour complexity blindly, experiments are designed around specific, AI-prioritised tissue interactions. - Back to patients (clinical relevance):
Predicted regulators and tissue programs are linked back to longitudinal patient cohorts, treatment response data, and survival outcomes, ensuring that discoveries are grounded in human disease and clinically meaningful.
Through this iterative cycle, AI-SEED-RCC reduces experimental trial-and-error, accelerates discovery, and enables the rational design of more accurate experimental models and therapeutic strategies.
Generating openly accessible foundational knowledge and datasets to power kidney cancer research
The collaboration’s tools, data, and results will be shared openly with the global research community. By decoding the “seed” from the “soil” of kidney cancer, the collaboration aims to:
- Generate rich information about the cells that drive untreated kidney cancer, helping to identify which patients need treatment and which can safely be monitored.
- Leverage information on the genetic and cellular features of a patient’s tumour to match the best available therapies for them.
- Support the development of new treatments that target the vasculature and immune cells that support tumours, to inhibit or reverse tumour growth.
Openly available spatial datasets paired with new generative AI models will also provide a foundation for future virtual tissue modelling across cancer types.
Diversity of expertise, experience and technologies
The AI-SEED-RCC collaboration is co-led by Dr Mo Lotfollahi and Dr Thomas Mitchell. It draws together clinicians, computational and experimental biologists and genomicists from across the Wellcome Sanger Institute, Cambridge Biomedical Campus and University College London. Collectively, their expertise encompasses:
- spatial biology
- kidney cancer genomics
- clinical oncology
- artificial intelligence
- generative artificial intelligence and machine learning model development
- translational immunology.
Outputs
By combining the above expertise with data from extensively characterised clinical cohorts of patients with untreated clear cell renal cell carcinoma (ccRCC), and data from patients profiled before and after treatment with VEGFR-directed tyrosine kinase inhibitors (TKIs) and immune checkpoint inhibitors (ICIs), we seek to generate the most comprehensive spatially resolved dataset of ccRCC to date.
The openly-available data resource will integrate:
- spatial transcriptomics
- proteomics
- in situ DNA sequencing
- immune receptor profiling
- clinical metadata
- 3D imaging of tissues.
These datasets will directly support generative AI model development, virtual tissue perturbation, and in silico testing of therapeutic strategies.
Sanger people

Prof Mats Nilsson
Professor of Biochemistry at Stockholm University and Associate Faculty at the Sanger Institute

Dr Mo Lotfollahi
Group Leader

Prof Muzlifah Haniffa
Head of Cellular Genomics, Senior Group Leader and Deputy Director of the Wellcome Sanger Institute

Dr Omer Bayraktar
Group Leader
External Contributors

Dr Thomas Mitchell
Project Lead - University Cambridge

Professor Grant Stewart
Project Specialist - University of Cambridge

Dr James Jones
Project co-lead (UK), University of Cambridge

Dr Mireia Crispin-Ortuzar
Project co-lead (UK), University of Cambridge

Professor David Long
Project co-lead (UK), University College London

Professor Maxine Tran
Project Specialist, University College London