CRISPRi + scRNA-seq + HIPSCI
Large-scale CRISPRi screens with scRNA-seq read-out in iPSCs
Genome-scale single-cell CRISPR in iPSCs from multiple individuals
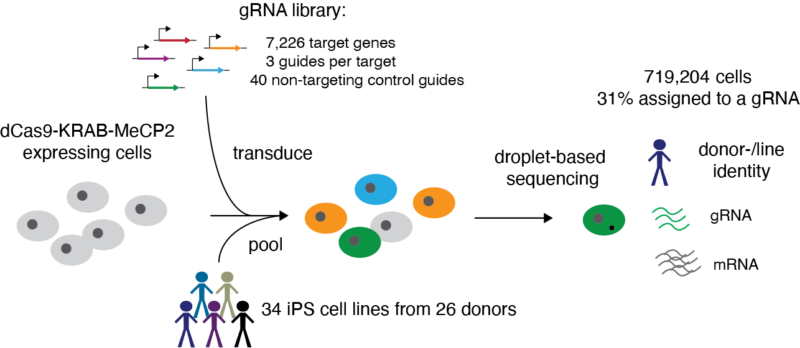
In the first portion of the paper, we apply CRISPRi to reduce expression on 7,226 genes in 34 iPSC lines from 26 donors to determine the mean effect of perturbation.
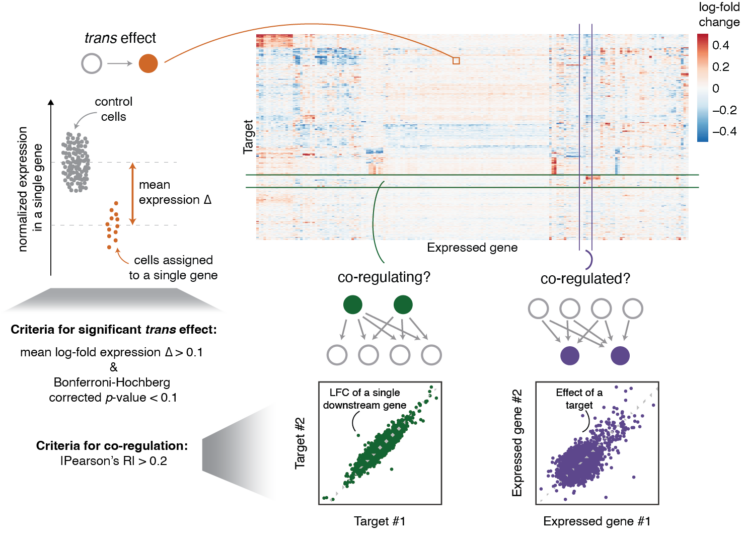
Our analysis strategy is roughly broken down into three aspects: i) trans effect of a target gene on downstream genes reflects links in a regulatory network (orange), (ii) correlation of trans effects for two target genes indicates similar function of the targets (green), and (iii) correlation of gene responses in trans indicates common control by shared regulators (purple).
For more, check out our interactive data browser and our pre-print!
Targeted single-cell CRISPR in iPSCs from multiple individuals
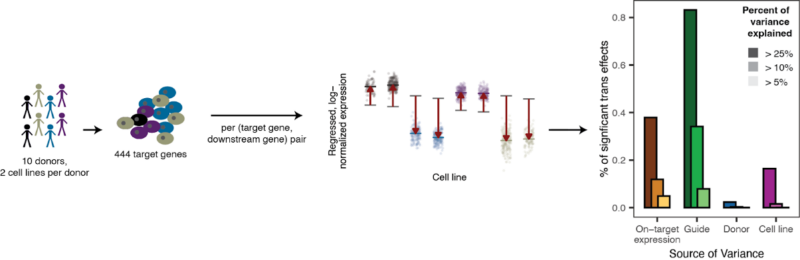
In the latter portion of the paper, we conduct another single-cell CRISPR screen on a more targeted panel of genes in 20 cell lines derived from 10 donors (with 2 cell lines per donor) to gain an understanding of the technical and biological factors driving variation of effect due to perturbation. Here, we proritized genes with variable roles in different biological contexts, such as heritably expressed genes, eQTL hotspots and genes associated with monogenic disease.
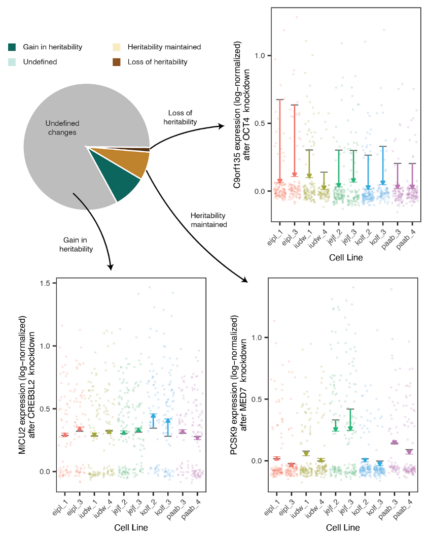
Of those effects driven by genetic variation, trans effects were roughly categorized into four groups based on the heritability of their wild-type expression levels. If the gene expression is heritable in wild-type, heritable trans effects can emerge due to near-complete repression, or other outcomes that remove the variation between donors (loss of heritability). Alternatively, expression heritability can be preserved where transcriptional change amplified effects of known genetic regulators like cis eQTLs or was small compared to the natural variation between donors (maintenance of heritability). In a second scenario, we considered heritability upon knockdown of the target genes to be potentially gained, if we could not find evidence for heritable expression in the control cells (gain in heritability).
For more, check out our interactive data browser [if forced to reload, too many users right now] and our pre-print!
Data access
Raw sequencing files (.fastq and .cram) are available from SRA under the accession number ERP16533 at https://www.ebi.ac.uk/ena/browser/view/PRJEB81502. LFC tables and a table describing the 516 sequencing files are available on FigShare at https://www.doi.org/10.6084/m9.figshare.26819743 and count data at https://www.doi.org/10.6084/m9.figshare.27989294. Additional results and processed data are available as Supporting Material for the publication.
Code availability
Code used for processing and analysis of the data is available on Github.
Socials
Check out our tweetorials on LinkedIn, X and BlueSky.
Contact
Questions? Get in touch with Claudia, Britta, Leo or Oli! We’re excited that you’re interested in our work!
Sanger Institute Contributors

Vivek Iyer
Human Genetics Informatics Team Lead

Dr Leopold Parts
Group Leader

Dr Oliver Stegle
Associate Faculty in the Cellular Genomics Programme
Previous contributors

Britta Velten
Visiting Scientist
External Contributors

Britta Velten
Principal Investigator

Jana Braunger
PhD Student