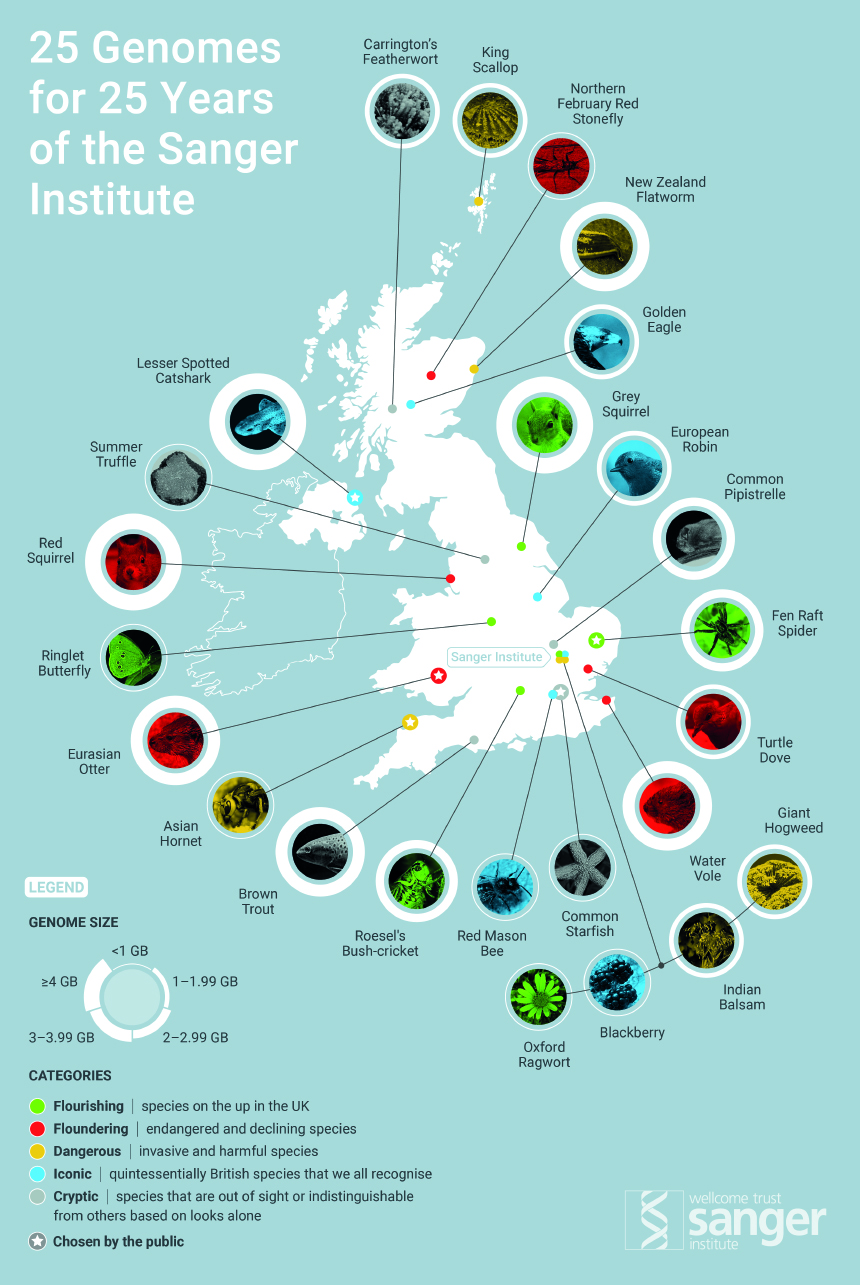
25 UK species' genomes sequenced for first time
The genomes of 25 UK species* have been read for the first time by scientists at the Wellcome Sanger Institute and their collaborators**. The 25 completed genome sequences, announced today (4 October) on the Sanger Institute’s 25th anniversary, will lead to future studies to understand the biodiversity of the UK and aid the conservation and understanding of our species.
The newly-sequenced genomes will enable research into why some brown trout migrate to the open ocean, whilst others don’t, or investigations into the magneto receptors in robins’ eyes that allow them to ‘see’ the magnetic fields of the Earth. The genomes could also help to shed light on why red squirrels are vulnerable to the squirrel pox virus, yet grey squirrels can carry and spread the virus without becoming ill.
The Sanger Institute was founded in 1993 by Professor Sir John Sulston as part of the Human Genome Project. The Institute made the largest single contribution*** to the gold- standard sequence of the first human genome, which was published in 2003.
A genome is an organism’s complete set of genetic instructions written in DNA. Each genome contains all of the information needed to build that organism and allow it to grow and develop.
Since the landmark completion of the human genome, the Sanger Institute has become a globally recognised leader in the field of genomics. Many more important reference genomes have already been sequenced – from the mouse and zebrafish genomes to the pig, gorilla, mosquito and many others. Beyond animal species, infectious diseases and bacteria also feature prominently on the list of reference genomes, from salmonella and MRSA to chlamydia and malaria. All of these have offered up important insights about these species in health and disease.
Now, the Sanger Institute and its partners have comprehensively sequenced 25 UK species for the first time. The first human genome took 13 years and billions of dollars to complete. With the great advances in technology and falling costs of sequencing, scientists have been able to newly sequence 25 species’ genomes in less than one year and at a fraction of the cost.
“We are thrilled to announce the completion of 25 genomes from UK species. Sequencing these species for the first time didn’t come without challenges, but our scientists and staff repeatedly came up with innovative solutions to overcome them. We have learned much through this project already and this new knowledge is flowing into many areas of our large scale science. Now that the genomes have been read, the pieces of each species puzzle need to be put back together during genome assembly before they are made available.”
Dr Julia Wilson Associate Director of the Sanger Institute
“We are already discovering the surprising secrets these species hold in their genomes. We’ve found that King scallops are more genetically diverse than we are, and the Roesel’s bush cricket’s genome is four times the size of the human genome. Similar to when the Human Genome Project first began, we don’t know where these findings could take us.”
Dan Mead Co-ordinator of Sanger’s 25 Genomes Project
The 25 Genomes Project has been made possible by PacBio® long-read sequencing technology, which generates high-quality genomes. The Institute partnered with PacBio and other leaders in the technology sector, 10x Genomics and Illumina, to create the most comprehensive view of these genomes.
The high-quality genomes will be made freely available to scientists to use in their research. Researchers could discover how UK species are responding to environmental pressures, and what secrets they hold in their genetics that enable them to flourish, or flounder.
“The 25 Genomes Project has uncovered the blueprints of the diversity of UK life, which will effectively rewrite what we know about these species. By comparing those blueprints within and between species we can understand the genetic diversity of fauna and flora from the UK and beyond. These newly-sequenced genomes are a starting point that will reveal aspects of evolution we’ve not even dreamt of.”
Dr Tim Littlewood Head of Life Sciences at the Natural History Museum, London and a partner of the 25 Genomes Project
This project is a small contribution to a much larger undertaking, where scientists from around the world are coming together to form a plan to sequence all life on Earth.
“DNA sequencing technology has advanced over the last number of years to a point at which we can at least discuss the possibility of sequencing the genomes of all of life on Earth. From those DNA sequences we will obtain inestimable insights into how evolution has worked, and to how life has worked.”
Professor Sir Mike Stratton Director of the Sanger Institute
More information
*The 25 species that have had their genomes sequenced are:
Flourishing species:
- Grey Squirrel
- Ringlet Butterfly
- Roesel’s Bush-Cricket
- Oxford Ragwort
Floundering species:
- Red Squirrel
- Water Vole
- Turtle Dove
- Northern February Red Stonefly
Dangerous species:
- Giant Hogweed
- Indian Balsam
- King Scallop, also known as Great Scallop, Coquilles Saint-Jacques
- New Zealand Flatworm
Iconic species:
- Golden Eagle
- Blackberry
- European Robin
- Red Mason Bee
Cryptic species:
- Brown trout
- Common Pipistrelle Bat
- Carrington’s Featherwort
- Summer truffle
Five species chosen by the public:
- Common Starfish
- Fen Raft Spider
- Lesser Spotted Catshark
- Asian Hornet
- Eurasian Otter
For more information on the 25 Genomes Project
Visit: https://www.sanger.ac.uk/collaboration/25-genomes-25-years
Our partners
**The 25 Genomes Project is a collaborative effort involving many institutions. Our partners include:
- Natural History Museum, London
- EMBL-EBI
- Pacific Biosciences (PacBio)
- The National Trust
- The Wildlife Trust
- Nottingham Trent University
- Edinburgh University
- 10x Genomics
- Illumina
- Aberdeen Centre for Environmental Sustainability (ACES)
- Animal and Plant Health Agency
- Buglife
- Cardiff University
- Centre for Agriculture and Biosciences (CAB) International
- Centre for Ecology and Hydrology
- Centre for Environment, Fisheries and Aquaculture Science (CEFAS)
- Ecole Polytechnique federale de Lausanne
- Institute for Marine Research, Norway
- James Hutton Institute
- Mycorrhizal Systems Ltd
- Natural History Museum of Geneva
- NC State University
- NIAB EMR
- Norwegian University of Life Sciences
- Observatoire Océanologique, Banyuls
- Open Air Laboratories (OPAL)
- Orthoptera & Allied Insects
- Oxford University
- Personal Chair of Zoological and Conservation Medicine
- Queen Mary’s University
- Queens University Belfast
- Royal Botanical Garden Edinburgh
- Royal Society for the Protection of Birds (RSPB)
- University College Dublin
- University College London
- University of Aberdeen
- University of Arkansas
- University of Bristol
- University of Edinburgh
- University of Exeter
- University of Lincoln
- University of Liverpool
- University of Nottingham
- University of Reading
- USDA
- Wellcome Genome Campus Grounds Team
- Wellcome Genome Campus Public Engagement
- Wildwood Trust
- www.dolomedes.org.uk
The Human Genome Project
***Of the 23 human pairs of chromosomes, eight were sequenced by researchers at the Sanger Institute and their collaborators.
Funding:
This project is supported by Wellcome.
Selected websites
About Pacific Biosciences
Pacific Biosciences of California, Inc. (NASDAQ:PACB) offers sequencing systems to help scientists resolve genetically complex problems. Based on its novel Single Molecule, Real-Time (SMRT®) technology, Pacific Biosciences’ products enable: de novo genome assembly to finish genomes in order to more fully identify, annotate and decipher genomic structures; full-length transcript analysis to improve annotations in reference genomes, characterize alternatively spliced isoforms in important gene families, and find novel genes; targeted sequencing to more comprehensively characterize genetic variations; and real-time kinetic information for epigenome characterization. Pacific Biosciences’ technology provides high accuracy, ultra-long reads, uniform coverage, and the ability to simultaneously detect epigenetic changes. PacBio® sequencing systems, including consumables and software, provide a simple, fast, end-to-end workflow for SMRT Sequencing. More information is available at www.pacb.com.
Natural History Museum
The Natural History Museum exists to inspire a love of the natural world and unlock answers to the big issues facing humanity and the planet. It is a world-leading science research centre, and through its unique collection and unrivalled expertise it is tackling issues such as food security, eradicating diseases and managing resource scarcity. The Natural History Museum is the most visited natural history museum in Europe and the top science attraction in the UK; we welcome more than 4.5 million visitors each year and our website receives over 500,000 unique visitors a month. People come from around the world to enjoy our galleries and events and engage both in-person and online with our science and educational activities through innovative programmes and citizen science projects. www.nhm.ac.uk
About 10x Genomics
10x Genomics is a leading company in the field of genomics. The company was founded on the vision that this century will bring unprecedented advances in biomedicine to transform the way we understand and treat diseases leading to dramatic improvements in human health. 10x products enable the acceleration of genetic discoveries through unparalleled resolution leading to a new understanding of biology and diseases. The company’s customers include leading biomedical research institutes, pharmaceutical companies and clinical centers. Founded in 2012, 10x Genomics is financed by marquee global investors including Venrock, Softbank, Foresite Capital, Fidelity, and Meritech Capital. For more information, visit www.10xgenomics.com.
Illumina
Illumina is improving human health by unlocking the power of the genome. Our focus on innovation has established us as the global leader in DNA sequencing and array-based technologies, serving customers in the research, clinical, and applied markets. Our products are used for applications in the life sciences, oncology, reproductive health, agriculture, and other emerging segments. To learn more, visit www.illumina.com and follow @illumina.
European Bioinformatics Institute (EMBL-EBI)
The European Bioinformatics Institute (EMBL-EBI) is a global leader in the storage, analysis and dissemination of large biological datasets. EMBL-EBI helps scientists realise the potential of ‘big data’ by enhancing their ability to exploit complex information to make discoveries that benefit humankind. EMBL-EBI is at the forefront of computational biology research, with work spanning sequence analysis methods, multi-dimensional statistical analysis and data-driven biological discovery, from plant biology to mammalian development and disease. We are part of the European Molecular Biology Laboratory (EMBL), an international, innovative and interdisciplinary research organisation funded by 22 member states and two associate member states, and are located on the Wellcome Genome Campus, one of the world’s largest concentrations of scientific and technical expertise in genomics. www.ebi.ac.uk
About Wellcome Genome Campus Public Engagement
Through a wide range of projects, activities, visits and events Wellcome Genome Campus Public Engagement encourages exploration and discussion about genomics, from exciting research findings to the social and ethical questions it can raise. Working together with collaborators in scientific research, the arts and humanities, public engagement and education specialists, and cultural organisations, we aim to share knowledge, spark discussions, and foster a community of engaged researchers. Wellcome Genome Campus Public Engagement is part of Connecting Science. www.wellcomegenomecampus.org/publicengagement
The Wellcome Sanger Institute
The Wellcome Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease. To celebrate its 25th year in 2018, the Institute is sequencing 25 new genomes of species in the UK. Find out more at www.sanger.ac.uk or follow @sangerinstitute on Twitter, Facebook and LinkedIn
Wellcome
Wellcome exists to improve health for everyone by helping great ideas to thrive. We’re a global charitable foundation, both politically and financially independent. We support scientists and researchers, take on big problems, fuel imaginations and spark debate. wellcome.org





