The ANOSPP Project
Aims of the ANOSPP Project
 The primary aim of the ANOSPP project is to improve our understanding of Anopheles species diversity, population structure, and malaria transmission across Africa. We will achieve this in collaboration with partners working in malaria endemic countries. 500,000 Anopheles mosquitoes collected from 100 locations across Africa over the next five years will undergo targeted sequencing with the ANOSPP amplicon panel. The sequencing data that results from each individual mosquito reveals its species, population structure, and whether Plasmodium parasites are present – key information for vector control endeavours.
The primary aim of the ANOSPP project is to improve our understanding of Anopheles species diversity, population structure, and malaria transmission across Africa. We will achieve this in collaboration with partners working in malaria endemic countries. 500,000 Anopheles mosquitoes collected from 100 locations across Africa over the next five years will undergo targeted sequencing with the ANOSPP amplicon panel. The sequencing data that results from each individual mosquito reveals its species, population structure, and whether Plasmodium parasites are present – key information for vector control endeavours.
While our Project’s focus is on African species, an important secondary goal is to complete a “reference index” for the entire Anopheles genus – this complete index is fundamental to the utility of ANOSPP. This requires mosquitoes or DNA extracts from at least 10 expertly identified individuals for every Anopheles species on the planet. For any expertly identified sample contributing to the reference index, we will also sequence COI and/or ITS2. All data are returned to partners and made openly available.
A key feature of this project is that we prefer to carry out the sequencing on DNA that has been non-destructively extracted so that our research partners may return to the specimen for further morphological examination when unexpected results arise. We are able to run the panel on whole mosquitoes, head/thoraxes, or even single legs if required.
About the ANOSPP Panel
 The ANOSPP panel is a multilocus amplicon sequencing panel that targets:
The ANOSPP panel is a multilocus amplicon sequencing panel that targets:
- 62 phylogenetically informative loci (each approx. 160bp long) in a generic Anopheles genome
- 2 conserved loci in a generic Plasmodium mitochondrion.
The panel uses a highly multiplexed two-step PCR that first amplifies target sequences and then adds unique barcodes, making it possible to pool amplicons from hundreds of mosquitoes into a single Illumina MiSeq sequencing run (Makunin et al. 2022).
Sequences are de-multiplexed using these barcodes and analysed using k-mer approaches to assign a species identity for each individual (Boddé et al. 2022).
We run population genetic analyses with the resulting mosquito data and we identify Plasmodium presence and species for any mosquitoes determined to be carrying parasites.
The ANOSPP Workflow
 Using our cheap, high-throughput, non-destructive DNA extraction workflow, where mosquito morphology can be studied after sequencing-based identification, we examine a very small aliquot of this DNA with a simple and fast targeted sequencing panel that works on any species from the Anopheles genus. Our workflow massively reduces the effort, time, and cost of carrying out confident species identification.
Using our cheap, high-throughput, non-destructive DNA extraction workflow, where mosquito morphology can be studied after sequencing-based identification, we examine a very small aliquot of this DNA with a simple and fast targeted sequencing panel that works on any species from the Anopheles genus. Our workflow massively reduces the effort, time, and cost of carrying out confident species identification.
Once DNA is sequenced with this panel, the resulting data reveal:
- the species identity
- geographic population structure within species
- if Plasmodium parasites are present in the mosquito
- the species of Plasmodium carried by positive mosquitoes
Therefore, the tool is like a Swiss army knife:
- multifunctional
- cheap
- easy to use
Benefits of using the ANOSPP panel
The ANOSPP panel has a number of important benefits for malaria researchers:
- ANOSPP is more accurate and informative than any other species identification technique and can be used on any Anopheles sample (adult, larva, eDNA sample from breeding water, etc.).
- Our non-destructive DNA extraction workflow supports the preservation of specimens for morphological study after sequencing, and specimens can be returned to partners for further study.
- All sequence data are generated at Sanger using Sanger funds, so partner costs are restricted to those incurred for sample collection and, if possible, shipment.
- Data are generated from ~800 individual mosquitoes in a single MiSeq run, and partners receive all resulting data from their samples and online data analysis training videos.
- For partners that are primarily interested in the results, we provide an “ANOSPP Report Card”, revealing the species identity of each Anopheles individual and whether or not it carries Plasmodium parasites, and if so, of what species.
Interested in running ANOSPP on your samples?
Anyone can apply for a partner study, though we are particularly looking to collaborate with partners who:
- Are planning or actively carrying out a study in Africa that includes mosquito samples collected across a range of locations or using a variety of approaches for longitudinal monitoring.
- Have access to a wide range of specimens or DNA from confidently identified Anopheles species from anywhere in the world, including historic (e.g. pinned) specimens collected from the 1970s onwards.
Partner Study Timelines
To apply to run a Partner Study, the following process and timeline will be followed:
Register your Interest – Duration: less than 1 month
Fill in this survey to register your interest in participating in ANOSPP for your research project.
Onboarding – Duration: 3-12 months
Legal and ABS compliance, with support from Sanger plus onboarding training on sample and metadata collection.
Collect Samples – Duration: up to the partners
Complete your collections, fill in the sample manifest, and submit it for validation.
Send Samples to ANOSPP for sequencing and analysis – Duration: up to the partners
Validated sample sets including more than eight 96-well plates are ideal – ship to the Sanger Institute for extractions and sequencing.
Reports and Data delivered to partners by ANOSPP: Duration 3-6 months
Receive a ‘ANOSPP report’ on your sample set as well as all sequencing data.
ANOSPP Data and Results
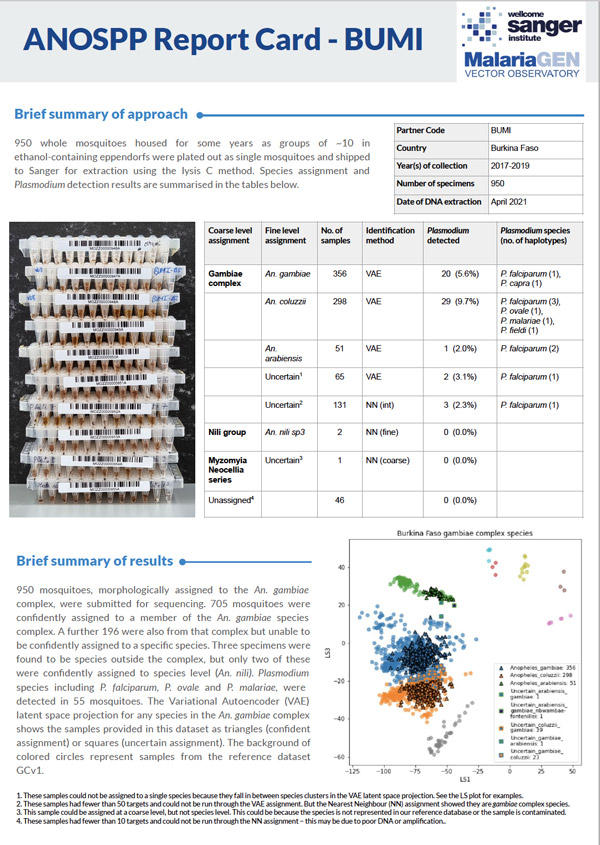 The sequencing data and all results are made available to our partners as soon as possible, and these are also released publicly within 12 months.
The sequencing data and all results are made available to our partners as soon as possible, and these are also released publicly within 12 months.
We provide a species ID for all mosquito samples with sufficient sequencing data and if this is not possible, indicate the species complex or group most similar to the sample.
We also provide quality-controlled haplotype data for each amplified locus, which can be used for further population genomic analyses.
Additionally, for every specimen we report whether Plasmodium was detected and if so, the species ID.
All the above results, combined with metadata captured in the partner manifest, are used to generate the ANOSPP Report (see right).
The data output that we share with our partners and eventually release publicly consists of a combined results table, a haplotype table, and the ANOSPP Report. See github.com/mariloubodde/NNoVAE for an example of the data output.
Contact the ANOSPP Project
To register your interest in using ANOSPP or contributing to the project, please go to tinyurl.com/ANOSPP
For further information on the application process or on the ANOSPP panel, contact anospp@sanger.ac.uk
Sanger people

Dr Mara Lawniczak
Senior Group Leader

Dr Petra Korlevic
Staff Scientist

Dr Alex Makunin
Computational Staff Scientist
Previous Sanger people

Marilou Boddé
Postdoctoral Fellow
External Contributors

Diego Ayala
Institut de Recherche pour le Développement, MIVEGEC, Univ. Montpellier, France
Affiliated Sites
External