More than 100 new gut bacteria discovered in human microbiome
Scientists working on the gut microbiome* have discovered and isolated more than 100 completely new species of bacteria from healthy people’s intestines. The study from the Wellcome Sanger Institute, Hudson Institute of Medical Research, Australia, and EMBL’s European Bioinformatics Institute, has created the most comprehensive collection of human intestinal bacteria to date. This will help researchers worldwide to investigate how our microbiome keeps us healthy, and its role in disease.
Reported today (4 February) in Nature Biotechnology, the new resource will allow scientists to detect which bacteria are present in the human gut, more accurately and faster than ever before. This will also provide the foundation to develop new ways of treating diseases such as gastrointestinal disorders, infections and immune conditions.
About 2 per cent of a person’s body weight is due to bacteria and the intestinal microbiome is a major bacterial site and an essential contributor to human health. Imbalances in our gut microbiome can contribute to diseases and complex conditions such as Inflammatory Bowel Disease, Irritable Bowel Syndrome allergies and obesity. However, as many species of gut bacteria are extremely difficult to grow in the laboratory, there is a huge gap in our knowledge of them.
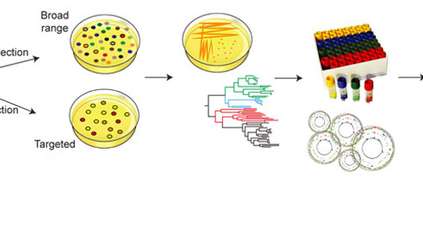
In this study, researchers studied faecal samples from 20 people from the UK and Canada, and successfully grew and DNA sequenced 737 individual bacterial strains from these. Analysis of these isolates revealed 273 separate bacterial species, including 173 that had never previously been sequenced. Of these, 105 species had never even been isolated before.
“This study has led to the creation of the largest and most comprehensive public database of human health-associated intestinal bacteria. The gut microbiome plays a major role in health and disease. This important resource will fundamentally change the way researchers study the microbiome.”
Dr Samuel Forster First author on the paper from the Wellcome Sanger Institute and Hudson Institute of Medical Research, Australia
Standard methods to understand how the gut microbiome impacts on human health involves sequencing the DNA from mixed samples of gut bacteria to try to understand each component. However, these studies have been severely hampered by the lack of individually isolated bacteria and reference genomes from them.
The new culture collection and reference genomes will make it much cheaper and easier for researchers to determine which bacteria are present within communities of people and research their role in disease.
“For researchers trying to find out which species of bacteria are present in a person’s microbiome, the database of reference genomes from pure isolates of gut bacteria is crucial. Then if they want to test a hypothesis, for example that a particular species is enriched in a certain disease, they can get the isolate itself from the collection and physically test in the laboratory if this species seems to be important.”
Dr Rob Finn An author from EMBL’s European Bioinformatics Institute
“This culture collection of individual bacteria will be a game-changer for basic and translational microbiome research. By culturing the unculturable, we have created a resource that will make microbiome analysis faster, cheaper and more accurate and will allow further study of their biology and functions. Ultimately, this will lead us towards developing new diagnostics and treatments for diseases such as gastrointestinal disorders, infections and immune conditions.”
Dr Trevor Lawley Senior author from the Wellcome Sanger Institute
More information
Publication:
Samuel C. Forster & Nitin Kumar et al. (2019) Human Gastrointestinal Bacteria Genome and Culture Collection. Nature Biotechnology. DOI: 10.1038/s41587-018-0009-7
* There are trillions of microbes in the human intestine, which form a huge ecosystem known as the gut microbiome. These microbes are known to be important for health, and imbalances in the gut microbiome can contribute to complex conditions and diseases.
Funding:
This work was supported by Wellcome, the Medical Research Council, the BBSRC, the European Molecular Biology Laboratory, the Australian National Health and Medical Research Council and the Victorian Government.
Selected websites
Hudson Institute of Medical Research
Hudson Institute is a leading Australian medical research institute recognised internationally for research into cancer, inflammation, reproduction and children’s health. Our research spans discovery research, translational research and clinical trials – progressing scientific knowledge into new an innovative treatments and cures.
Hudson Institute is part of a major scientific and research medical innovation cluster in Melbourne, Australia. With our precinct partners, Monash Health and Monash University, our research is informed by clinical challenges and developed into clinics.
Our precinct’s Translational Research Facility accelerates the results of medical research by bringing together leading scientists and clinicians supported by state-of-the-art technologies clinical trials centre. A link bridge connects the facility to the Monash Medical Centre, providing a crucial flow between scientific discovery and health outcomes. More: https://hudson.org.au, Twitter, Facebook, Linkedin
EMBL’s European Bioinformatics Institute (EMBL-EBI)
EMBL’s European Bioinformatics Institute (EMBL-EBI) is a global leader in the storage, analysis and dissemination of large biological datasets. EMBL-EBI helps scientists realise the potential of ‘big data’ by enhancing their ability to exploit complex information to make discoveries that benefit humankind.
EMBL-EBI is at the forefront of computational biology research, with work spanning sequence analysis methods, multi-dimensional statistical analysis and data-driven biological discovery, from plant biology to mammalian development and disease.
EMBL-EBI is part of the European Molecular Biology Laboratory (EMBL), an international, innovative and interdisciplinary research organisation funded by 25 member states and two associate member states, and are located on the Wellcome Genome Campus, one of the world’s largest concentrations of scientific and technical expertise in genomics. www.ebi.ac.uk
Wellcome Sanger Institute
The Wellcome Sanger Institute is one of the world’s leading genome centres.
Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
Find out more at www.sanger.ac.uk or follow @sangerinstitute on Twitter, Facebook, LinkedIn and on our Blog.
About Wellcome
Wellcome exists to improve health by helping great ideas to thrive.
We support researchers, we take on big health challenges, we campaign for better science, and we help everyone get involved with science and health research.
We are a politically and financially independent foundation. wellcome.org