Bryant Group
Lung microbiome
We apply cutting-edge approaches to study the genetic makeup and cellular activity at work in the microbiome and patients’ lungs. In addition, we use single-cell profiling and advanced computational analyses to gain insights that we hope will lead to future diagnostics and treatments for chronic lung conditions.
Related Sanger Institute Blog:
Breath of fresh air - interview with Josie Bryant
Introduction
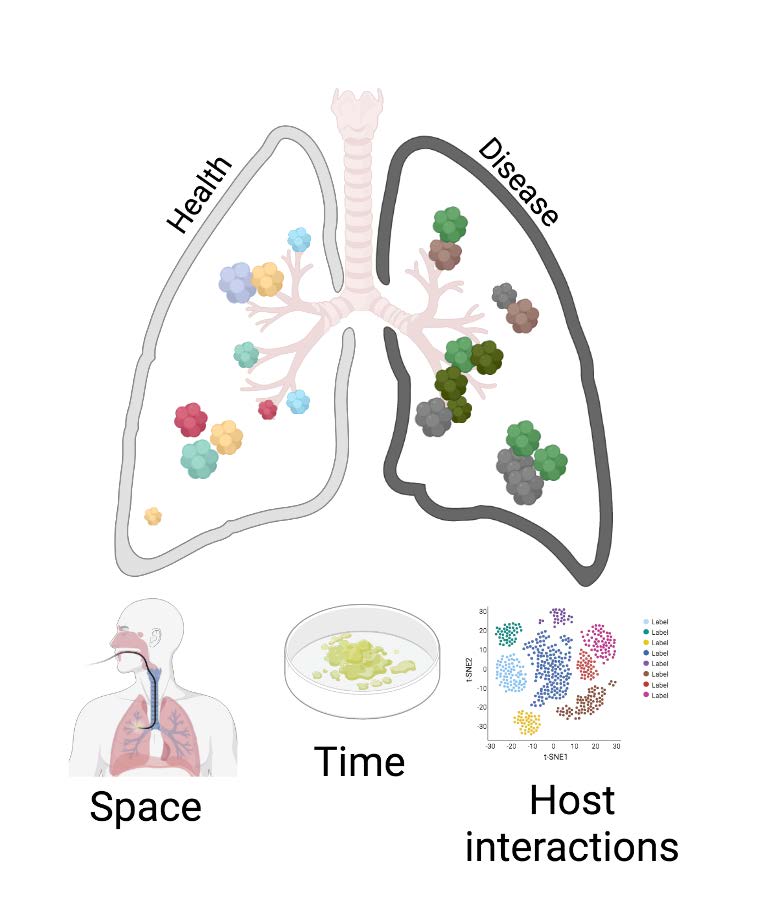
(a) in different parts of the lungs;
(b) over time.
The team also studies microbiome-host
interactions through the use of metabolomics and single-cell transcriptomics.
Our lungs are exposed to, and colonized, by a wide range viruses, bacteria and fungi, which collectively form our respiratory microbiome. Increasing evidence suggests that the respiratory microbiome has a vital role in the normal development of the immune system, as well as maintaining health over a lifetime by protecting against acute bacterial and viral infections.
In people with chronic lung conditions, changes in this complex microbial community can have serious effects on diseases progression. However, at present we have relatively little understanding of how changes in the respiratory microbiome are associated with lung damage, or how the microbiome contributes to normal lung health. This lack of knowledge limits our ability to develop new microbial-based diagnostics and treatment approaches for chronic lung diseases.
To fill in these blanks, we collaborate with colleagues in Addenbrookes and Papworth Hospitals to couple cutting-edge genomic, metabolomic and proteomic approaches with advances in sampling techniques, single-cell profiling and advanced computational analyses to study the respiratory microbiome in unprecedented detail.
Tracking the changes in the respiratory microbiome over time in both healthy people and those with chronic lung diseases will enable us to determine:
- the bacteria, viruses, and fungi that are associated with long-term lung conditions
- how specific strains and mutations within each species in the respiratory microbiome contribute to health and disease
- the interplay and communication between the respiratory microbiome and a person’s lung cells in normal and damaged lungs.
We aim to use these insights to identify biomarkers of health and disease, and uncover pathways of interaction between the microbiome and host cells, which can provide opportunities to develop new diagnostic tests and treatment targets.
Goals
Our team aims to provide the research community with:
- a multi-dimensional understanding of respiratory microbiome dynamics in health and disease
- a list of potential biomarkers of lung disease progression and therapeutic targets for treatment of chronic respiratory conditions
- new droplet-based whole-genome multiple displacement amplification (WG-MDA) methods for identifying and analysing microbes in low biomass samples.
Research approach
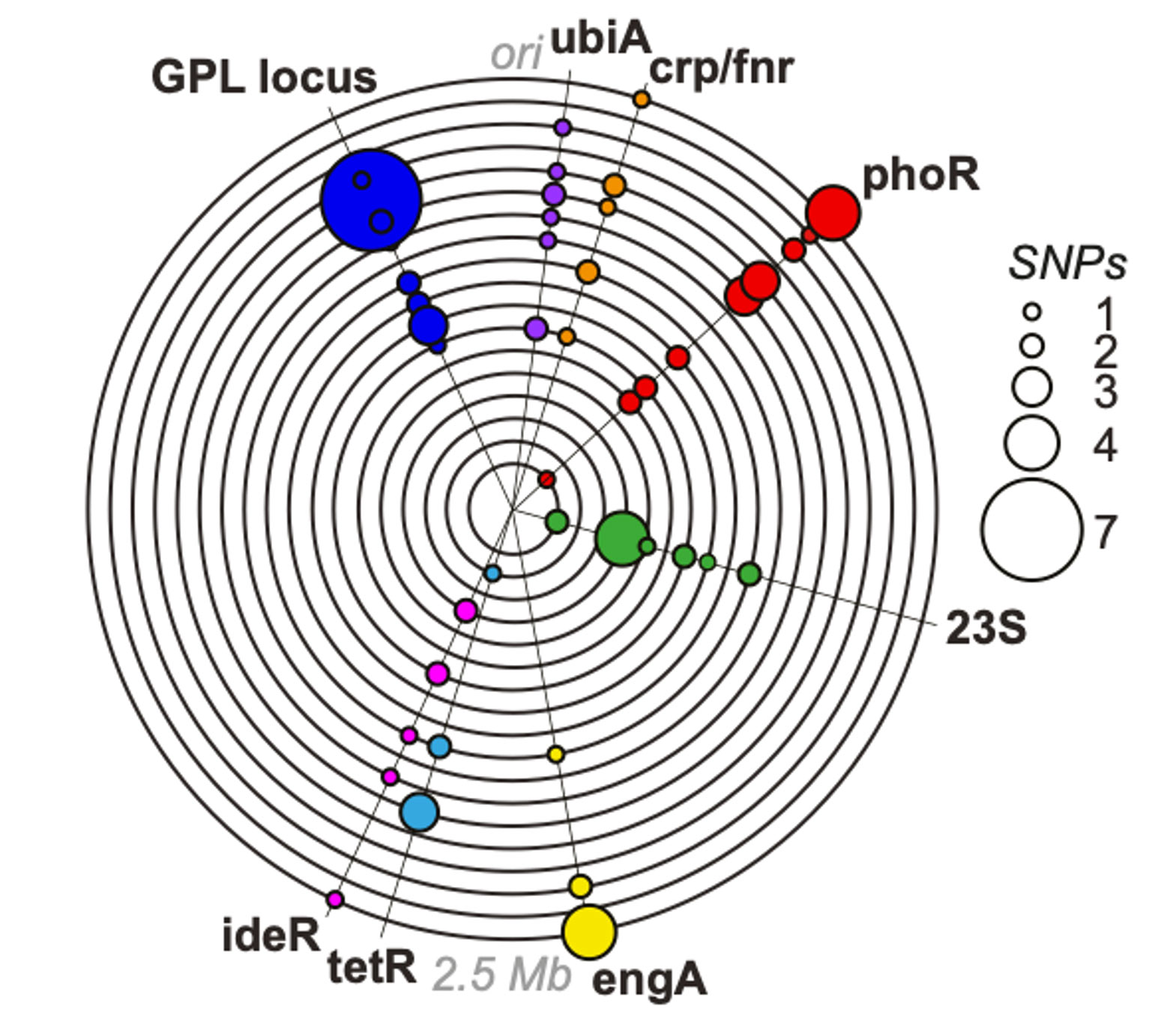
(Bryant et al 2021 Science)
Working closely with our clinical collaborators, the team combines advanced airway sampling and dense longitudinal collection with state of the art ‘omic’ approaches to accurately describe the ecology of the respiratory microbiome in both space and time, and define causal relationships to lung pathobiology.
We characterise inter- and intra- species diversity (across bacteria, fungi and viruses) in health and disease using metagenomics. This enables us to identify changes in the microbiome in chronic lung conditions that are associated with, and potentially causing, clinical deteriorations such as pulmonary exacerbations.
In parallel, we use metabolomics, proteomics and single-cell transcriptomics to explore the interactions between the microbiome and host cells. In this way, we are building a multi-dimensional understanding of the lung niche in both health and disease.
Join our team
The team is always on the lookout for bright and motivated people to join my group so, if you’re interested, please send Josie an email and include your CV.
Core team

Dr Seri Kitada
PhD Student

Cecilia Kyanya
PhD Student

Javier Magide Marco
PhD Student

Dr Duy Truong Pham
Senior Computer Biologist

Dr Belén Saavedra
Postdoctoral Fellow
Related groups
Partners
We work with the following groups
