The inside story of England COVID pandemic described in new study
The COVID-19 crisis that gripped the UK between September 2020 and June 2021 can be thought of as a series of overlapping epidemics, rather than a single event, say researchers at the Wellcome Sanger Institute, EMBL’s European Bioinformatics Institute (EMBL-EBI) and their collaborators. During this period, the country wrestled with several versions of the SARS-CoV-2 virus that possessed different biological properties and required a different public health response.
The study, published today (14 October) in Nature, is the most detailed analysis of SARS-CoV-2 genomic surveillance information to date. It describes the scientific ‘story’ of the pandemic as it unfolded and underlines the importance of high-speed, large-scale genomic surveillance to understand and respond to infectious outbreaks.
In March 2020, just as the UK was preparing to enter the first of several lockdowns, the COVID-19 Genomics UK (COG-UK) consortium was set up to monitor the spread and evolution of SARS-CoV-2 by sequencing the virus’s genome1.
Since then, the consortium has identified and monitored numerous viral variants, including the Alpha variant first identified in Kent in September 2020 and the Delta variant first identified in India in April 2021. Both of these variants subsequently changed the course of the pandemic, not only in the UK but globally.
For this study, researchers at the Wellcome Sanger Institute and EMBL-EBI analysed SARS-CoV-2 genomic surveillance data from England2 collected between September 2020 and June 2021. They characterised the growth rates and geographic spread of 71 lineages and reconstructed how newly emerging variants changed the course of the epidemic.
At the end of 2020, the Alpha variant (B.1.1.7) spread despite a series of restrictions, including a national lockdown in November and regional restrictions in December. Though these measures slowed the spread of other variants, Alpha was found to possess a 50 to 60 per cent growth advantage over previous variants and continued to spread rapidly.
In the system of tiered restrictions operating in December 2020, infection rates were higher in areas with fewer restrictions. Alpha was only brought under control in a third national lockdown between January and March 2021, which was introduced after a peak of 72,088 daily cases on 29 December. This measure simultaneously eliminated most variants that had been dominant in September and October 2020. When restrictions began to be lifted on 8 March 2021, the daily case rate had fallen to 5,500.
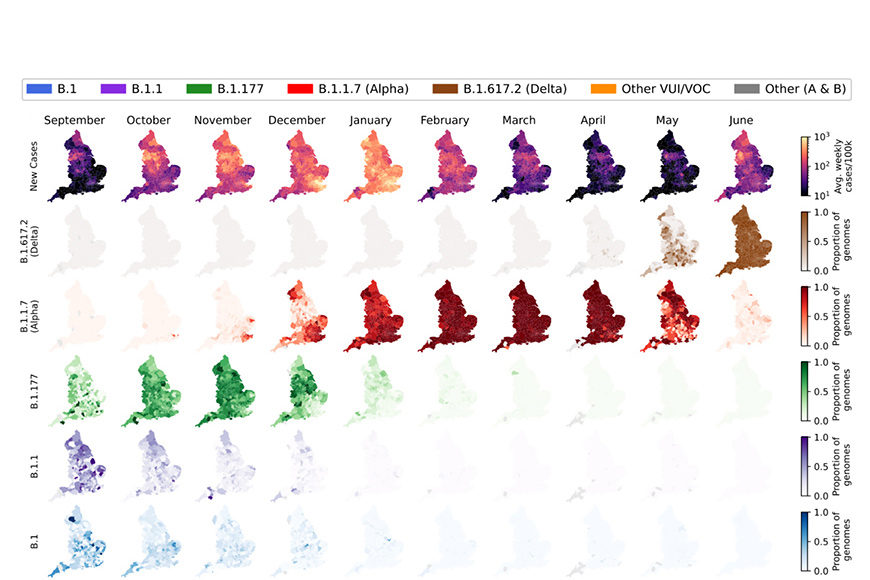
While Alpha was being brought under control, variants associated with a greater ability to circumvent immunity from vaccination or prior infection continued to appear in the UK at low levels in early 2021. These variants were characterised by the spike mutation E484K, the most significant of which were the Beta variant (B.1.351, first identified in South Africa) and Gamma variant (P.1, first identified in Brazil). But despite repeated introductions of these variants, they were confined to short-lived local outbreaks.
In March 2021, the first samples of B.1.617, which originated in India, began appearing in sequence data. This was in fact two lineages, Kappa (B.1.617.1) and Delta (B.1.617.2). Though Delta contained different mutations to previous variants of concern, these mutations achieved even greater transmissibility3. While Kappa grew slowly and has since faded away, Delta had spread to all local authorities and accounted for 98 per cent of viral genomes sequenced by 26 June 2021.
“Time has proven how ingenious an idea it was to set up the COVID-19 Genomics UK (COG-UK) consortium at the beginning of the pandemic. Being able to see lineages side-by-side, mapped to specific locations, has been incredibly informative in terms of understanding how this series of epidemics has unfolded. To see Alpha growing faster in nearly 250 out of 315 local authorities was a clear signal that we were dealing with something very different. At the same time, we’ve learned that the genetics of SARS-CoV-2 are incredibly complex. Even though we knew all of Delta’s mutations, it wasn’t immediately clear that it would become the dominant lineage, for example.”
Dr Moritz Gerstung, a senior author of the paper from EMBL-EBI and The German Cancer Research Centre (Deutsches Krebsforschungszentrum, DKFZ)
Analysis of Delta indicates that its growth rate was 59 per cent higher than that of Alpha, the greatest growth advantage observed in any other variant to date. Overall, the researchers estimate that the spread of more transmissible variants between August 2020 and the early summer of 2021 more than doubled the average growth rate of the virus in England.
“Thanks to genomic surveillance in the UK and internationally, it is clear that we are dealing a virus that has changed considerably since the one that we faced in March 2020. We will continue to monitor the SARS-CoV-2 virus to ensure that we can use the most effective vaccines, treatments and public health measures against current and future variants.”
Dr Meera Chand, COVID-19 incident director at the UK Health Security Agency (UKHSA) and one of the authors of the paper
Although it remains impossible to predict what the virus will do next, the COG-UK consortium has advanced the field of genomic surveillance considerably and proven the value of monitoring for infectious agents. Just 18 months after its inception, the programme catalysed the establishment of national sequencing systems that provide near real-time epidemiological information to inform the UK’s public health response. It is hoped that one day scientists will be able to predict the emergence of new variants.
“These genomic surveillance data have given us a totally new way of watching an outbreak unfold, which has taught us a lot about how a new infectious agent spreads and evolves. My hope is that similar genomic surveillance programmes will be developed across the world, so that we are as well-prepared as we can be to respond to future infectious disease outbreaks – whether they be familiar pathogens or new ones.”
Dr Jeff Barrett, a senior author of the paper and Director of the COVID-19 Genomics Initiative at the Wellcome Sanger Institute
More information
1 An overview of how COVID-19 genomes are sequenced is available on the Sanger Institute blog. More information on COG-UK is available on the COG-UK website.
2 These data track SARS-CoV-2 lineages in 315 Lower Tier Local Authorities (LTLAs) in England. An LTLA is an administrative region with approximately 100,000–200,000 inhabitants. In total, 281,178 viral genomes were sequenced during this period.
3 Kappa and Delta contained the L452R spike protein mutation, thought to reduce antibody recognition, and P681R, which helps the virus enter human cells in a similar manner to Alpha’s P681H mutation. Delta contains 5 additional spike protein mutations, the consequences of which are not fully understood yet.
Publication:
Harald S. Vöhringer, Theo Sanderson and Matthew Sinnott et al. (2021). Genomic reconstruction of the SARS-CoV-2 epidemic in England. Nature. DOI: 10.1038/s41586-021-04069-y
Funding:
COG-UK is supported by funding from the Medical Research Council (MRC) part of UK Research & Innovation (UKRI), the National Institute of Health Research (NIHR) and Genome Research Limited, operating as the Wellcome Sanger Institute.