Study reveals how immune cells target different tissues
For the first time, researchers have revealed the different molecular identities of important immune cells, called T regulatory cells using single cell genomics, in both mouse and human peripheral non-lymphoid tissues such as skin and colon. The researchers from the Wellcome Sanger Institute and their collaborators revealed that T regulatory cells have tissue-specific receptors and other adaptations, which allow them to move to and remain in the correct location in the body. In future, this could allow us to understand how to target therapeutic cells to specific places in the body, for targeted treatments of autoimmune diseases for example.
Reported in Immunity (5th February), the study showed that mouse T regulatory cells had multiple intermediate cell states, within and between tissues. They also discovered similar patterns of gene activities in humans, revealing more detail than ever about how the immune system is regulated.
T regulatory cells (Tregs) are a specialised type of immune cell that control the immune system. They dampen down the immune response to keep it in check and prevent the body attacking its own tissues. These cells are found in lymphoid tissue such as lymph nodes and spleen, and also in other non-lymphoid tissues in the body. While some differences are known about how T regulatory cells function in different tissues, understanding the “GPS system” that addresses cells to specific tissue in the body is limited.
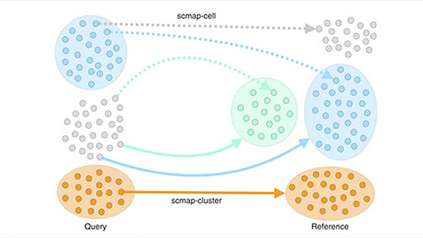
To create a detailed picture of T cells, the researchers studied 35,000 individual cells from healthy mouse lymph nodes, spleen, skin and colon, comparing cells across the different tissues. They used single-cell RNA sequencing to discover exactly which genes were switched on in each of the cells. The researchers discovered that different tissues had different populations of cells, and revealed the changes that T regulatory cells undergo when moving between tissues.
Further analysis revealed that even within a single tissue, cells varied greatly – with a continuum of cell states and regulatory activities. They also discovered that although the sequence of cell states is very similar when they migrate to different peripheral tissues such as skin and colon, cells express distinct chemokine ‘location’ receptors on their surface that act as a satnav to guide them to a specific tissues.
“This is the first time that anyone has described the huge varied spectrum of T regulatory cell populations in peripheral tissues. We can see that although these cells share the core identity of T regulatory cells, they are very different across different tissues, with different functions, and even express different receptors to guide them to a specific tissue. This is helping us understand the regulation of the immune system to keep it in a healthy balance.”
Tomas Gomes Joint first author from the Wellcome Sanger Institute
The researchers analysed Tregs from mouse tumour and non-tumour cells in specific tissues and saw they had the same gene activities. Comparing the mouse data with Tregs from human skin, colon and blood tissues they discovered similar receptors and signalling molecules in humans..
““We now know that Tregs play unique roles in different tissues, some of which are distinct from their suppressive function. By revealing the common and distinct molecular signals that support different Treg populations, our study allows a better understanding of how Tregs interact with their environment to promote tissue health throughout the body.”
Professor Fiona Powrie From the University of Oxford
“This is the most comprehensive study ever performed of single cell RNA sequencing of T regulatory cells across tissues. Not only does it help us understand the immune system within a tissue, it also reveals which regulators and receptors are expressed in each tissue. This could help researchers learn how to manipulate potential therapeutic T cells in the future, to design them for specific locations in the body and target exactly the right tissue needed.”
Dr Sarah Teichmann Corresponding author from the Wellcome Sanger Institute
More information
Publication:
Ricardo J Miragaia and Tomas Gomes et al. (2019). Single Cell transcriptomics of regulatory T cells reveals trajectories of tissue adaptation. Immunity. DOI: 10.1016/j.immuni.2019.01.001
Funding:
This work was supported by the ERC, EMBO, the Fundacao para Ciencia e Tecnolgia, Portugal, the EU’s Horizon 2020 research and innovation programme Enlight-Ten, and a Marie Curie fellowship.
Selected websites
Wellcome Sanger Institute
The Wellcome Sanger Institute is one of the world’s leading genome centres.
Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
Find out more at www.sanger.ac.uk or follow @sangerinstitute on Twitter, Facebook, LinkedIn and on our Blog.
About Wellcome
Wellcome exists to improve health by helping great ideas to thrive.
We support researchers, we take on big health challenges, we campaign for better science, and we help everyone get involved with science and health research.
We are a politically and financially independent foundation. wellcome.org