DECIPHER - Mapping the Clinical Genome
DECIPHER is an interactive web-based database which incorporates a suite of tools designed to aid the interpretation of genomic variants. DECIPHER enhances clinical diagnosis by retrieving information from a variety of bioinformatics resources relevant to the variant found in the patient. The patient’s variant is displayed in the context of both normal variation and pathogenic variation reported at that locus thereby facilitating interpretation.
Archive Page
This page is maintained as a historical record and is no longer being updated.
The DECIPHER database and team moved to EMBL’s European Bioinformatics Institute (EMBL-EBI) in July 2023. The database can be accessed at: https://www.deciphergenomics.org/
About
DECIPHER is a web-based platform for secure deposition, analysis, and sharing of plausibly pathogenic genomic variants from well-phenotyped patients suffering from rare genetic disorders. DECIPHER aids clinical interpretation of these rare sequence and copy-number variants by providing tools for variant analysis and identification of other patients exhibiting similar genotype–phenotype characteristics. DECIPHER also provides mechanisms to encourage collaboration among a global community of clinical centers and researchers, as well as exchange of information between clinicians and researchers within a consortium, to accelerate discovery and diagnosis. DECIPHER has contributed to matchmaking efforts by enabling the global clinical genetics community to identify many previously undiagnosed syndromes and new disease genes, and has facilitated the publication of over 800 peer-reviewed scientific publications since 2004.
Contributing to the DECIPHER database is an international community of academic departments of clinical genetics and rare disease genomics now numbering more than 250 projects and having uploaded more than 19,000 cases. Each contributing centre has a nominated rare disease clinician or clinical geneticist who is responsible for overseeing data entry and membership for their centre. DECIPHER enables a flexible approach to data-sharing. Each project maintains control of its own patient data (which are password protected within the project’s own DECIPHER project) until consent is given to share the data with chosen parties in a collaborative group or to allow anonymous genomic and phenotypic data to become freely viewable. Once data are shared, consortium members are able to gain access to the patient report and contact each other to discuss patients of mutual interest.
Home page
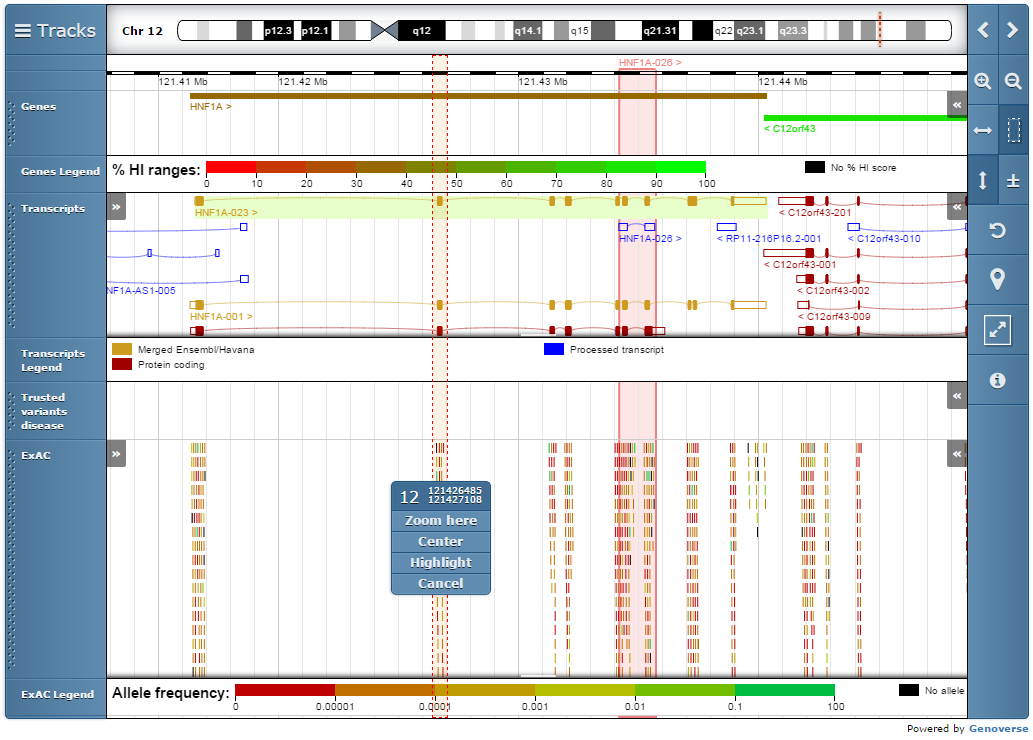
Genoverse viewer
Downloads
Please access DECIPHER from the following website: https://www.deciphergenomics.org/
Further information
The DECIPHER consortium provides these data in good faith as a research tool, but without verifying the accuracy, clinical validity or utility of the data. The DECIPHER consortium, makes no warranty, express or implied, nor assumes any legal liability or responsibility for any purpose for which the data are used.
For licensing options, please see: https://www.deciphergenomics.org/datasharing.
Please cite DECIPHER with the following publication:
DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources. Firth, H.V. et al (2009). Am.J.Hum.Genet 84, 524-533 (DOI: dx.doi.org/10/1016/j.ajhg.2009.03.010)
Contact
If you need help or have any queries, please contact us using the details below.
We are always eager for feedback on DECIPHER and any suggestions for improvement. Please write to us at contact@deciphergenomics.org.
Sanger Institute Contributors

Dr Paul Bevan
Head of Core Software Services

Professor Matthew Hurles
Director the Wellcome Sanger Institute and Senior Group Leader