
Dr Paul Bevan, MBCS
Head of Core Software Services
As head of Core Software Services I lead a team that supports the three Wellcome funded entities on campus, namely the Wellcome Sanger Institute; the Wellcome Genome Campus and; Wellcome Connecting Science. I also play a role as Customer Relationship Manager in supporting the Biodata Innovation Centre (BIC); also funded by Wellcome and hosted on campus. My team comprises the following services: Core Web; Core Bioinformatics (CoreBio); and Core Web Security. These services are provided to all Wellcome funded activities on the Wellcome Genome Campus.
The Wellcome Sanger Institute is a world-leading genomics research institute. Our work helps improve human health and understand life on Earth. Underpinning much of this work is a reliance on the Institute’s Informatics and Digital Solutions (IDS) presence and web technologies to store, interpret and make available genomic research data sets to the scientific community world-wide. My team ensures the Wellcome Sanger Institute research data are made available through the use of web technologies to the external scientific community and the public in a timely fashion in accordance with the Institute’s strategic plans and funding.
My team likewise supports the Wellcome Genome Campus itself, for which we provide its web presence and on campus bespoke IT requirements. Wellcome Connecting Science, the programme which supports teaching, learning; courses and conferences as well as on Campus venue facilities, is also provided with its web presence and bespoke IT requirements by the team.
From a technology perspective we use popular, well supported web frameworks to deliver requirements for the five research programmes based at the Wellcome Sanger Institute. We work closely with other teams in IDS: Database administrators (DBAs); Infrastructure Management; Informatics Support Group; Data Centre; Pipelines Group; Service Delivery; Service Desk. These are all important collaborations within our department of 140 people and allows IDS to respond rapidly to create tools and resources e.g. supporting the COVID sequencing resource that was set up within a month of the pandemic starting.
All of this is delivered at scale to provide the large data sets required for research in a timely and cost effective manner. Further details about our technical approach to providing web services can be found on our group page.
Another important aspect of the team’s remit is to ensure the web security for all the various enterprise, consortium and research group websites (of which there are 160 domains along with a further 180 subdomains). It is important that we keep our huge IT infrastructure safe and secure from hacking and attacks by malicious groups. We use proactive monitoring, risk assessment and agile development strategies to improve and secure as well as innovate our existing web services and to accommodate any ‘bigger picture’ requirements of the Institute on an on-going basis.
My prior background as a Postdoctoral Fellow research scientist provides the appropriate insight and understanding of working in the scientific research environment. I worked in the Departments of Medical Genetics and Clinical Biochemistry at the University of Cambridge and Department of Medicine at McGill University in Montreal Canada. My two major research interests were: Receptor Tyrosine Kinase Signal Transduction and; Autosomal Dominant Polycystic Kidney Disease.
During my time at the University of Cambridge I was webmaster for the Departments of Medical Genetics and Clinical Biochemistry as well as webmaster of the Cambridge Institute for Medical Research website.
My timeline
Transfer of the Decipher project to the EBI - https://www.deciphergenomics.org/
Additional role as Deputy head of Web security at the Wellcome Sanger Institute
Additional role as Customer Relationship Manager for the Biodata Innovation Centre
Member of the British Computer Society
Start of the Deciphering Developmental Disorders Study https://www.ddduk.org/
Head of Core Software Services at the Wellcome Sanger Institute
Head of the Core Web team at the Wellcome Sanger Institute
Start of the Decipher project https://www.deciphergenomics.org/
Joined the Wellcome Sanger Institute in Web and IT
Postdoctoral researcher in Department of Medical Genetics, University of Cambridge
Awarded a Wellcome Trust Research Project grant
Postdoctoral researcher in Department of Clinical Biochemistry, University of Cambridge
Awarded a Juvenile Diabetes Foundation International Postdoctoral Fellowship
Received the Renouf Award for outstanding achievement in Medical Research
Awarded a Royal Victoria Hospital Research Institute Postdoctoral Fellowship
Postdoctoral researcher at McGill University, Montreal, Canada
Awarded PhD in Medical Biochemistry from King's College, University of London
Received a Medical Research Council PhD Industrial Partnership Award
Awarded BSc in Applied Biochemistry from Liverpool John Moores University
Quick links
Programmes and Facilities
My publications

Receiving the award for Best Scientific website at the Laboratory News Industry Awards 2001, London. (For the Sanger Institute website - www.sanger.ac.uk.)

At the Wellcome building in Euston Road, London with the Book of life to whichthe Wellcome Sanger Institute made the largest sequencing contribution.
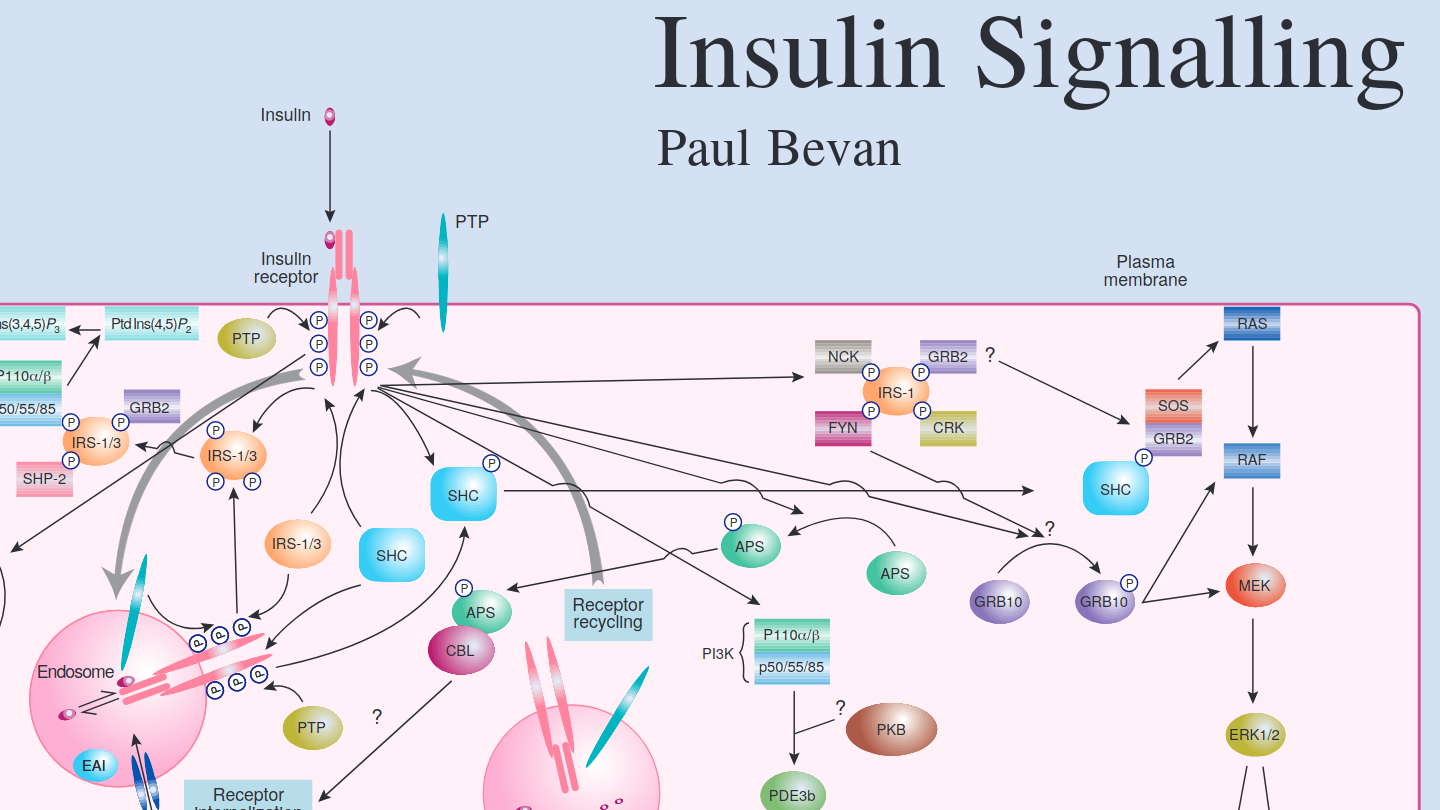
Insulin Signalling pathway schematic

Insulin receptor containing endosomes recycling

The annual international Decipher Symposium 2007 at Hinxton Hall on the Wellcome Genome Campus.

Hinxton Hall in the Snow 20101221

Hinxton Hall in the Snow 20101221

The Ensembl Web Team 20030401

Project team for Decipher to EBI 20230706
