Analysis of COVID-19 Genomes reveals large numbers of introductions to the UK in March
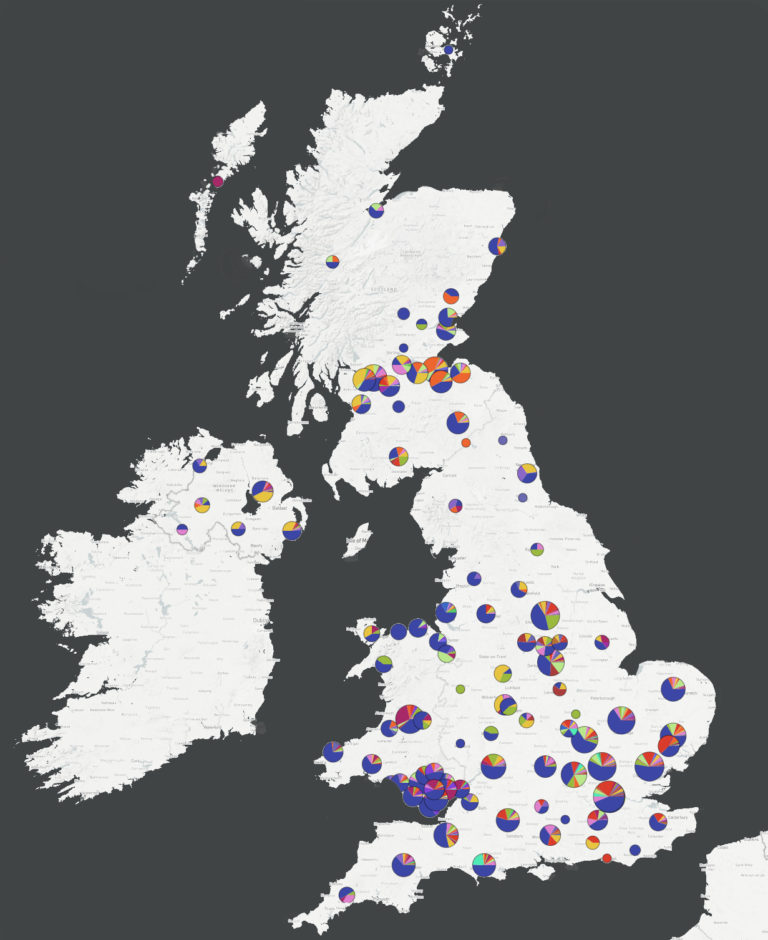
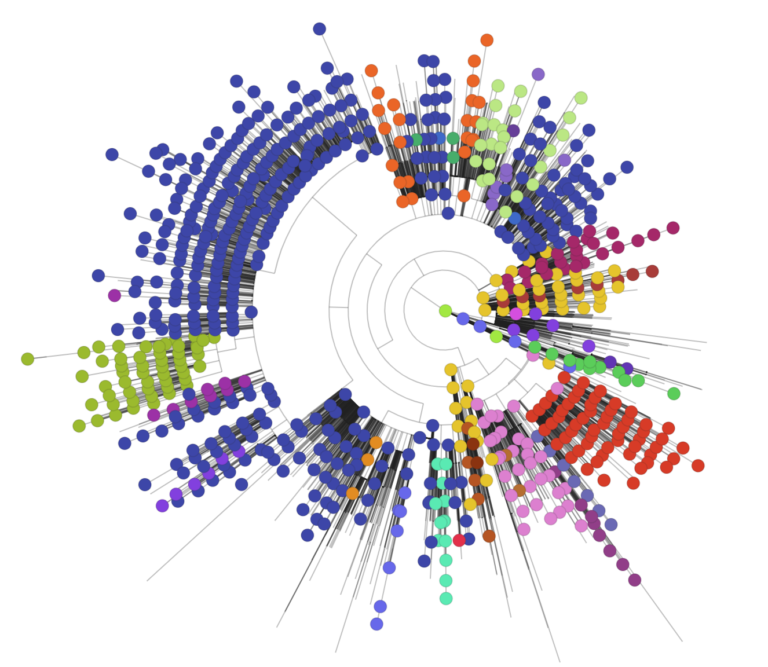
Approximately 40 lineages of the SARS-CoV-2 virus have been recorded circulating in the UK, some of which have already gone extinct while others thrive. Previously, the different lineages of the single strain of the virus entered the UK through multiple importations from around the globe, including European countries such as Spain, Italy and France.
Now, fewer international lineages remain in the UK and new cases of COVID-19 arise from local spread rather than importation from other countries. The data reports§, published by the COVID-19 Genomics UK Consortium (COG-UK), show the value in mapping COVID-19 lineages across the UK to understand how the virus is spreading at national, regional and local levels.
As of 22 May, more than 20,000 viral genomes from positive COVID-19 tests have been sequenced in the UK, which is the largest number of COVID-19 genomes sequenced by any single country affected by the pandemic.
These genome sequences are being generated and utilised by researchers within COG-UK to identify different lineages* of the virus circulating in the UK. Despite recent reports, this volume of sequencing confirms the all COVID-19 cases in the UK share a recent common ancestor from China; all cases are closely related. The lineages identified highlight small changes to the virus that enable monitoring and tracking over time, but do not signal the emergence of new strains at this point in the outbreak.
These data will aid the UK government in understanding patterns of spread in the UK to help focus different interventions in particular areas of the UK to control the spread of the virus and ultimately save lives.
The COVID-19 Genomics UK Consortium (COG-UK) – comprised of the NHS, Public Health Agencies and numerous academic and research institutions – is delivering large scale, rapid sequencing of the SARS-CoV-2 virus from positive COVID-19 samples and sharing intelligence with hospitals, regional NHS centres and the Government.
The latest COG-UK data reports show large numbers of independent introductions of the virus into the UK from around the world, resulting in approximately 40 COVID-19 lineages that are currently, or have been, circulating in the UK. However, recent data suggest new COVID-19 cases in the UK are arising from local spread rather than people travelling to the UK from other countries.
A dynamic lineage assignment method**, developed within the consortium, enables monitoring of SARS-Cov2 dynamics over time within the UK, with data mapped and delivered openly through an interactive web application***.
The data generated through COG-UK will be used to provide virus status reports, including estimates for the reproduction number, at the level of cities or local authorities.
“By analysing and comparing the different lineages of the SARS-CoV-2 virus that causes COVID-19 we can see how the virus is spreading through the UK, and determine whether new cases are arising from local spread versus importation from other countries. By ongoing mapping of lineages in space and time we can better understand and react to the changing dynamics of the pandemic.”
Professor David Aanensen, Director of the Centre for Genomic Pathogen Surveillance at the University of Oxford Big Data Institute
“Since starting this project in late March, this consortium has sequenced more than 20,000 viral genomes from positive COVID-19 samples. Undertaking a project of this scale is made possible by an amazing network of collaborators and our staff who have applied their world-class expertise in genomics and surveillance of infectious diseases to tackle COVID-19.”
Dr Cordelia Langford, Director of Scientific Operations at the Wellcome Sanger Institute
“This virus is one of the biggest threats our nation has faced in recent times and crucial to helping us fight it is understand how it is spreading. Harnessing innovative genome technologies will help us tease apart the complex picture of coronavirus spread in the UK, and rapidly evaluate ways to reduce the impact of this disease on our society.”
Professor Sharon Peacock, Director of the COVID-19 Genomics UK Consortium (COG-UK)
More information
* What is a viral lineage?
When the virus replicates, genetic errors – mutations – happen in the virus’ genome. When comparing viruses, those with shared patterns of mutations are assigned to different lineages. However a new lineage is not the same as a new strain. A new strain must differ from related viruses in significant ways, such as their transmissibility or virulence. Viral lineages can be plotted on a phylogenetic tree, with the branches representing different lineages, to show the evolutionary relationships between the various lineages. This provides insight into how the virus is mutating and spreading.
** http://pangolin.cog-uk.io and https://www.biorxiv.org/content/10.1101/2020.04.17.046086v1
*** https://microreact.org/project/cogconsortium
Publication:
COG-UK data reports for weeks 2 (1 April) and 3 (9 April) are available here: https://www.cogconsortium.uk/news/
Funding:
COG-UK is supported by £20 million funding from the UK Department of Health and Social Care (DHSC), UK Research and Innovation (UKRI) and the Wellcome Trust, administered by UK Research and Innovation.
Selected websites
COVID-19 Genomics UK Consortium (COG-UK)
The current COVID-19 pandemic, caused by the SARS-CoV-2 virus, represents a major threat to health. The COVID-19 Genomics UK (COG-UK) consortium has been created to deliver large-scale and rapid whole-genome virus sequencing to local NHS centres and the UK government.
COG-UK is made up of an innovative partnership of NHS organisations, the four Public Health Agencies of the UK, the Wellcome Sanger Institute and over twelve academic partners providing sequencing and analysis capacity. A full list of collaborators can be found here: https://www.cogconsortium.uk/about/
COG-UK is supported by £20 million funding from the UK Department of Health and Social Care (DHSC), UK Research and Innovation (UKRI) and the Wellcome Trust, administered by UK Research and Innovation. For more information, visit: https://www.cogconsortium.uk/
The Centre for Genomic Pathogen Surveillance
The Centre for Genomic Pathogen Surveillance is an initiative based at the Big Data Institute, University of Oxford and co-housed at the Wellcome Sanger Institute focussed on genomic epidemiology, laboratory and software engineering for global surveillance of microbial pathogens. The Centre seeks to enable the provision of genomic and epidemiological big data and tools to allow researchers, doctors and governments worldwide to track and analyse the spread of pathogens and antimicrobial resistance. https://www.pathogensurveillance.net/
About the Big Data Institute
The Big Data Institute is located in the Li Ka Shing Centre for Health Informatics and Discovery at the University of Oxford. It is an interdisciplinary research centre that focuses on the analysis of large, complex data sets for research into the causes, consequences, prevention and treatment of disease. Research is conducted in areas such as genomics, population health, infectious disease surveillance and the development of new analytic methods. The Big Data Institute is supported by funding from the Medical Research Council, the Engineering, Physical Sciences Research Council, the UK Research Partnership Investment Fund, the National Institute for Health Research Oxford Biomedical Research Centre, Wellcome and philanthropic donations from the Li Ka Shing and Robertson Foundations. Further details are available at www.bdi.ox.ac.uk
About UK Research and Innovation
UK Research and Innovation works in partnership with universities, research organisations, businesses, charities, and government to create the best possible environment for research and innovation to flourish. It aims to maximise the contribution of each of our component parts, working individually and collectively. It works with many partners to benefit everyone through knowledge, talent and ideas. Now the UK has left the European Union, UKRI continues to support the research and innovation communities with information and updates on access to grants and mobility. Operating across the whole of the UK with a combined budget of more than £7 billion, UK Research and Innovation brings together the seven research councils, Innovate UK and Research England. https://www.ukri.org/
The Wellcome Sanger Institute
The Wellcome Sanger Institute is a world leading genomics research centre. We undertake large-scale research that forms the foundations of knowledge in biology and medicine. We are open and collaborative; our data, results, tools and technologies are shared across the globe to advance science. Our ambition is vast – we take on projects that are not possible anywhere else. We use the power of genome sequencing to understand and harness the information in DNA. Funded by Wellcome, we have the freedom and support to push the boundaries of genomics. Our findings are used to improve health and to understand life on Earth. Find out more at scion-02.sandbox.sanger.ac.uk or follow us on Twitter, Facebook, LinkedIn and on our Blog.
About Wellcome
Wellcome exists to improve health by helping great ideas to thrive. We support researchers, we take on big health challenges, we campaign for better science, and we help everyone get involved with science and health research. We are a politically and financially independent foundation. wellcome.org




