Wellcome Sanger Institute sequences reference genomes of 3,000 dangerous bacteria
The genomes of more than 3,000 bacteria, including some of the world’s most dangerous, have been sequenced by researchers at the Wellcome Sanger Institute in collaboration with Pacific Biosciences (PacBio). Infecting tens of millions of people worldwide every year, these bacteria have been collected by the National Collection of Type Cultures (NCTC) and include deadly strains of plague, dysentery and cholera.
 By decoding the DNA, researchers will be able to better understand these diseases and how they become resistant to antibiotics. The publicly available genomic maps could also lead to the development of new diagnostic tests, vaccines or treatments.
By decoding the DNA, researchers will be able to better understand these diseases and how they become resistant to antibiotics. The publicly available genomic maps could also lead to the development of new diagnostic tests, vaccines or treatments.
Set up in 1920, the NCTC is the longest established collection of bacteria in the world. With more than 5,500 species of bacteria so far, the NCTC is also one of the world’s largest collections of clinically relevant bacteria. It is used extensively by researchers who are comparing historical and modern strains to advance global knowledge about the epidemiology, virulence, prevention and treatment of infectious diseases.
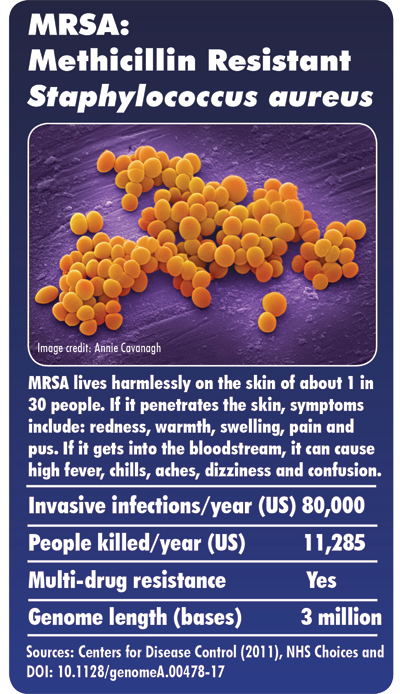
Antibiotic resistance is a significant problem globally and the collection includes some of the most important known drug-resistant bacteria. These include tuberculosis, one of the top ten causes of death worldwide, infecting 10.4 million and killing 1.7 million people in 2016 alone*, and gonorrhoea, the sexually transmitted disease that infects 78 million people a year** and is now becoming extremely difficult to treat. The NCTC also contains samples of methicillin-resistant Staphylococcus aureus (MRSA), which is resistant to multiple antibiotics and which can cause life-threatening infections in hospitals.
The genetic study of these strains will help researchers to understand the mechanisms of antibiotic resistance, and to look for any cracks in their armour to enable treatment.
 All ‘type strains’ of bacteria in the collection, the first strains that describe the species and are used to classify them, were sequenced as part of this initiative. The genome sequences of these highly valuable strains are fundamental for developing ways to identify specific infections in people, including tests diagnosing bacterial infections in the field to rapidly identify the source of an outbreak and help contain infections.
All ‘type strains’ of bacteria in the collection, the first strains that describe the species and are used to classify them, were sequenced as part of this initiative. The genome sequences of these highly valuable strains are fundamental for developing ways to identify specific infections in people, including tests diagnosing bacterial infections in the field to rapidly identify the source of an outbreak and help contain infections.
Amongst the many historically important strains in the collection are 16 deposited by penicillin discoverer Alexander Fleming, including a sample taken from his own nose. Also notable is the first bacteria to be deposited in the NCTC: A strain of dysentery-causing Shigella flexneri that was isolated in 1915 from a soldier in the trenches of World War 1***.
“Historical collections such at the NCTC are of enormous value in understanding current pathogens. Knowing very accurately what bacteria looked like before and during the introduction of antibiotics and vaccines, and comparing them to current strains from the same collection, shows us how they have responded to these treatments. This in turn helps us develop new antibiotics and vaccines. PacBio’s comprehensive DNA sequencing enables deep genomic analyses, and we are happy to be partnering with them for this important project.”
Dr Julian Parkhillfrom the Wellcome Sanger Institute
“The high-quality genomic maps enabled by SMRT® Sequencing allow an unprecedented understanding of these bacteria. We are delighted to be chosen by institutions like Wellcome Sanger Institute to help create such essential resources for the scientific and public health communities.”
Jonas Korlach Ph.D., Chief Scientific Officer of PacBio
“This resource is a vital tool for public health and by sequencing the bacteria, we have made the NCTC collection ready for the 21st century so that the research community can track and understand the bacteria. With this collection, we are providing tools for tracing infections, and identifying outbreaks of resistant bacteria, transforming public health in the UK.”
Dr Julie Russell Head of Culture Collections, which is operated by the National Infection Service of Public Health England
Going forward, all the bacterial species in the NCTC collection will be sequenced as they are collected. Researchers can order bacterial strains from the NCTC website at: https://www.phe-culturecollections.org.uk/collections/nctc.aspx. Full information about each strain, including the DNA sequences, are available at EMBL-EBI: https://www.ebi.ac.uk/ena/data/view/PRJEB6403.
More information
*WHO figures http://www.who.int/en/news-room/fact-sheets/detail/tuberculosis
** WHO figures Each year, an estimated 78 million people are infected with gonorrhoea*.
*** By sequencing a strain of dysentery-causing Shigella flexneri that was isolated in 1915 from a soldier in the trenches of World War 1 and comparing it to three other strains isolated in 1954, 1984 and 2002, researchers were able to determine an evolutionary pattern of the disease and learn about its virulence and antibiotic resistance.
https://www.sanger.ac.uk/news_item/2014-11-07-world-war-i-soldier-helps-in-fight-against-dysentery
https://www.youtube.com/watch?v=u-fbd9JpiMs
Funding:
This project was supported by Wellcome and Pacific Biosciences.
Forward-Looking Statements
All statements in this press release that are not historical are forward-looking statements, including, among other things, statements relating to the expected benefits of the collaboration between Pacific Biosciences and the Wellcome Sanger Institute, the suitability of products or technologies for particular applications, future availability, uses, quality or performance of, or benefits of using, products or technologies, and other future events. You should not place undue reliance on forward-looking statements because they involve known and unknown risks, uncertainties, changes in circumstances and other factors that are, in some cases, beyond Pacific Biosciences’ control and could cause actual results to differ materially from the information expressed or implied by forward-looking statements made in this press release. Factors that could materially affect actual results can be found in Pacific Biosciences’ most recent filings with the Securities and Exchange Commission, including Pacific Biosciences’ most recent reports on Forms 8-K, 10-K and 10-Q, and include those listed under the caption “Risk Factors.” Pacific Biosciences undertakes no obligation to revise or update information in this press release to reflect events or circumstances in the future, even if new information becomes available.
Selected websites
The National Collection of Type Cultures
NCTC is one of four culture collections that constitute the Culture Collections of Public Health England and has access to the wide-ranging expertise of an internationally-renowned body of specialists in clinical, food, water and environmental microbiology, genomics and proteomics. NCTC supplies reference bacterial cultures, including many type strains, of medical, scientific and veterinary importance worldwide. NCTC strains support academic, health, food and veterinary institutions and are used in microbiology laboratories in a range of different sectors and in research institutes worldwide. Founded in 1920, NCTC is the longest-established collection of its type anywhere in the world, and also serves as a United Nations Educational, Scientific and Cultural Organization (UNESCO) Microbial Resource Centre (MIRCEN). Find out more at www.phe-culturecollections.org.uk/nctc or follow us on Twitter @NCTC_3000
About Pacific Biosciences
Pacific Biosciences of California, Inc. (NASDAQ:PACB) offers sequencing systems to help scientists resolve genetically complex problems. Based on its novel Single Molecule, Real-Time (SMRT®) technology, Pacific Biosciences’ products enable: de novo genome assembly to finish genomes in order to more fully identify, annotate and decipher genomic structures; full-length transcript analysis to improve annotations in reference genomes, characterize alternatively spliced isoforms in important gene families, and find novel genes; targeted sequencing to more comprehensively characterize genetic variations; and real-time kinetic information for epigenome characterization. Pacific Biosciences’ technology provides high accuracy, ultra-long reads, uniform coverage, and the ability to simultaneously detect epigenetic changes. PacBio® sequencing systems, including consumables and software, provide a simple, fast, end-to-end workflow for SMRT Sequencing. More information is available at www.pacb.com.
The Wellcome Sanger Institute
The Wellcome Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease. To celebrate its 25th year in 2018, the Institute is sequencing 25 new genomes of species in the UK. Find out more at www.sanger.ac.uk or follow @sangerinstitute
Wellcome
Wellcome exists to improve health for everyone by helping great ideas to thrive. We’re a global charitable foundation, both politically and financially independent. We support scientists and researchers, take on big problems, fuel imaginations and spark debate. wellcome.org


