Machine learning flags emerging pathogens

A new machine learning tool that can detect whether emerging strains of the bacterium, Salmonella are more likely to cause dangerous bloodstream infections rather than food poisoning has been developed. The tool, created by a scientist at the Wellcome Sanger Institute and her collaborators at the University of Otago, New Zealand and the Helmholtz Institute for RNA-based Infection Research, a site of the Helmholtz Centre for Infection Research, Germany, greatly speeds up the process for identifying the genetic changes underlying new invasive types of Salmonella that are of public health concern.
Reported today (8 May) in PLOS Genetics, the machine learning tool could be useful for flagging dangerous bacteria before they cause an outbreak, from hospital wards to a global scale.
As the cost of genomic sequencing falls, scientists around the world are using genetics to better understand the bacteria causing infections, how diseases spread, how bacteria gain resistance to drugs, and which strains of bacteria may cause outbreaks.
However, current methods to identify the genetic adaptations in emerging strains of bacteria behind an outbreak are time-consuming and often involve manually comparing the new strain to an older reference collection.
The group of bacteria known as Salmonella includes many different types that vary in the severity of the disease they cause. Some types cause food poisoning, known as gastrointestinal Salmonella, whereas others cause severe disease by spreading beyond the gut, for example Salmonella Typhi which causes typhoid fever.
To understand the genetic changes that determine whether an emerging strain of Salmonella enterica will cause food poisoning versus a more severe infection, researchers built a machine learning model that analyses which mutations play an important role.
The team trained the model using old lineages of Salmonella that are evolutionarily distinct, including six Salmonella bacteria that caused invasive infections, and seven gastrointestinal strains of the bacteria. The machine learning model identified almost 200 genes involved in determining whether the bacterium will cause food poisoning or is better adapted to an invasive infection.
“We have designed a new machine learning model that can identify which emerging strains of bacteria could be a public health concern. Using this tool, we can tackle massive data sets and get results in seconds. Ultimately, this work will have a big impact on the surveillance of dangerous bacteria in a way we haven’t been able to before, not only in hospital wards, but at a global scale.”
Dr Nicole Wheeler Co-lead author from the Wellcome Sanger Institute
When applied to strains of Salmonella that are currently emerging in Sub-Saharan Africa, the tool correctly highlighted two types from a pool of commonly circulating infections (Salmonella Enteritidis and Salmonella Typhimurium) that are more dangerous and associated with higher numbers of bloodstream infection cases.
These infections are particularly bad in people with a weakened immune system, such as those with HIV. The machine learning tool revealed genetic changes that enabled Salmonella strains to adapt to their hosts and become more invasive.
“The machine learning tool is an advance compared to other methods as it not only searches for genes and mutations, it looks for the functional impacts mutations have in these bugs. It can tell us which mutations make pathogens better at spreading beyond the gut and causing a life-threatening disease rather than food poisoning. This will help in designing more effective treatments in the future.”
Dr Lars Barquist Co-lead author from the Helmholtz Institute for RNA-based Infection Research in Germany
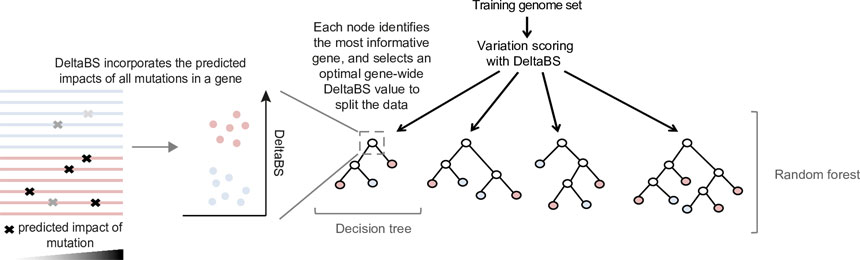
The machine learning tool, which produces an invasiveness index based on a random forest model*, is not limited to Salmonella and could be used to study other factors like emerging antibiotic resistance in any bacterium. It could be used in real time to identify a dangerous strain of bacteria before it spreads to cause an outbreak.
“We are already using this approach to look for key differences in strains of Salmonella Typhi circulating in Asia compared to Africa. Instead of manually comparing the genomes of different strains of bacteria over weeks or months, we are able to discover the genetic changes behind emerging strains of bacteria in seconds. It offers the potential to study outbreaks in real time and thus rapidly inform public health strategies to control or prevent disease.”
Dr Nicholas Feasey of the Liverpool School of Tropical Medicine
More information
*Random forests work by building an ensemble of decision trees designed to predict a characteristic of the samples, in this case the genetic changes behind the adaptation of bacteria to survive in an invasive or non-invasive environment (i.e. living within the bloodstream or in the gut).
Breiman L. Random Forests. Mach Learn. Kluwer Academic Publishers; 2001; 45: 5–32.
The invasiveness index ranks different types of bacteria on their predicted level of adaptation to invasive infection.
Publication:
Nicole Wheeler et al. (2018) Machine learning identifies signatures of host adaptation in the bacterial pathogen Salmonella enterica. PLOS Genetics. DOI: 10.1371/journal.pgen.1007333
Funding:
This work was supported by Wellcome (grant 206194), the University of Canterbury, the Biomolecular Interaction Centre, the Alexander von Humboldt Foundation, and a Rutherford Discovery Fellowship administered by the Royal Society of New Zealand.
Selected websites
Helmholtz Institute for RNA-based Infection Research
The Helmholtz Institute for RNA-based Infection Research (HIRI, http://www.helmholtz-hiri.de/) was established in May 2017 as a partnership between the Helmholtz Centre for Infection Research (HZI) and the Julius-Maximilian-University of Würzburg (JMU). HIRI is the first research institution worldwide bridging the fields of RNA biology and infectious disease.
Biomolecular Interaction Centre, University of Canterbury, New Zealand
The Biomolecular Interaction Centre (BIC, http://www.canterbury.ac.nz/bic/) is a multi-disciplinary research centre dedicated to the study of molecular interactions critical to biological function. Understanding these interactions is central to a range of fundamental sciences, new treatments for disease and a wide range of highly functional products.
The Wellcome Sanger Institute
The Wellcome Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease. To celebrate its 25th year in 2018, the Institute is sequencing 25 new genomes of species in the UK. Find out more at www.sanger.ac.uk or follow @sangerinstitute
Wellcome
Wellcome exists to improve health for everyone by helping great ideas to thrive. We’re a global charitable foundation, both politically and financially independent. We support scientists and researchers, take on big problems, fuel imaginations and spark debate. wellcome.org