Nodules, nitrogen and nature
 Researchers from the Universities of York, East Anglia and Reading, together with scientists from the Wellcome Trust Sanger Institute, have published their study of the genome of one of the remarkable bacterial species that support virtually all life on earth. Rhizobium leguminosarum and other species capture nitrogen from the atmosphere and convert it into forms that are usable by plants. Without this ‘fixation’ of nitrogen, life as we know it could not exist.
Researchers from the Universities of York, East Anglia and Reading, together with scientists from the Wellcome Trust Sanger Institute, have published their study of the genome of one of the remarkable bacterial species that support virtually all life on earth. Rhizobium leguminosarum and other species capture nitrogen from the atmosphere and convert it into forms that are usable by plants. Without this ‘fixation’ of nitrogen, life as we know it could not exist.
And while gardeners and farmers appreciate the value of these organisms, the study of rhizobia may also lead to advances in biology and medicine. Rhizobia are closely related to organisms that cause human disease, such as blood infections, and comparing the DNA of disease-causing organisms with the benign plant-dwelling Rhizobium should help scientists to explore disease processes in a harmless model.
The research also highlights a fundamental biological question: What makes a species?
The new report, together with other recent studies, highlights the observation that bacterial genomes have a large number of accessory genes that enhance a species’ ability to prosper in its environment, in addition to the core set of genes that carry out fundamental processes such as copying DNA and making cell walls.
But life is not simple.
“What we think of as defining of this organism – the ability to fix nitrogen – is not in its core set of genes or even on its chromosome: it is accessory and resides on a plasmid. Even more interesting, when we look at the closest relatives of Rhizobium leguminosarum, these include Agrobacterium tumefaciens, which doesn’t fix nitrogen and causes disease in plants.
“So these two species, closely related by some measures, have almost opposite consequences for plants, conferred by genes in the accessory set. Evolution in bacteria is proving to be a complex dance of genes among species.”
Professor Julian Parkhill Leader of the Pathogen Sequencing Unit at the Wellcome Trust Sanger Institute
For human biology, it is clear that virtually all our genes share a common inheritance back to and beyond divergence from the chicken. For bacteria, each gene or group of genes can have a different evolutionary history.
Most of the core genes of Rhizobium leguminosarum closely resemble those of Agrobacterium, but some are more similar to genes from other species. Even this most stable set of genes has a diverse history.
For the accessory genes, new analyses suggested that our views of bacterial evolution might be too simple.
“We looked at the composition of the accessory genes and, as in most species, they have a different genetic signature to the core genes. The common assumption is that they look different because they have been acquired more recently than the core set and so reflect, perhaps, the genetic signature of the organism from which they are derived.
“Our study shows this is not the case. The accessory genes look different for some other reason and we suspect it is because they are under different selective and mutation pressures to the core genes. The forces acting on accessory genes are different to those acting on core genes.”
Professor Peter Young Professor of Molecular Ecology at the University of York
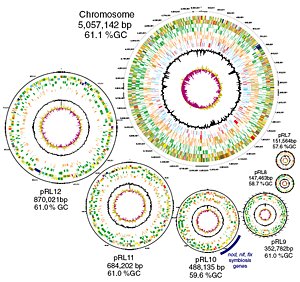
At 7.75 million bases of genetic code, the genome of Rhizobium leguminosarum is one of the largest bacterial genomes sequenced, and includes a large chromosome and six separate plasmids that resemble mini-chromosomes. The genome is so large that it has more genes than the more ‘complex’ budding yeast and more ‘chromosomes’ than fission yeast.
“Nearly 30 per cent of the genes in Rhizobium leguminosarum are not found in any other species – even with data for several other related organisms. With more than 7200 genes, there are new avenues to study the biology of this organism and its interaction with host plants.”
Dr Lisa Crossman Who led the annotation at the Wellcome Trust Sanger Institute
The soil is a diverse bacterial environment, with many species competing for resources and battling each other for survival. Rhizobium leguminosarum must survive between symbiosis opportunities among its vigorous competitors.
To survive and to prosper, it requires a diverse range of abilities and the ability to acquire new characteristics. Different members of the species have different numbers of plasmids – this strain has six that make up more than one-third of the genome.
Rhizobium leguminosarum is particularly important because it was the first rhizobial species to be described. The bacteria were first isolated in the 1890s by Beijerinck in the Netherlands.
Nitrogen-fixing bacteria in plant roots are essential to a healthy environment and leguminous plants can reduce reliance on chemical fertilisers. Genes involved in the symbiotic interactions with plants have been well studied, but its methods for survival and growth in the soil have not. The genome provides a new resource to understand those methods.
It is not simply health of the soil that might benefit from this work. Soil-dwelling bacteria share 2000 of 3300 predicted proteins with the species that causes the human and animal disease brucellosis. This species lives inside host cells and is thought to have evolved from a plant pathogen or symbiont.
“From each bacterial species we study, whether they infect plants or animals, we reveal more that is shared between them and more that helps inform both areas of research. The genome sequence of Rhizobium leguminosarum feeds directly into our understanding of the biology and evolution of organisms such as MRSA and Clostridium difficile.”
Professor Julian Parkhill Sanger Institute
More information
Publications:
Selected websites
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute, which receives the majority of its funding from the Wellcome Trust, was founded in 1992. The Institute is responsible for the completion of the sequence of approximately one-third of the human genome as well as genomes of model organisms and more than 90 pathogen genomes. In October 2006, new funding was awarded by the Wellcome Trust to exploit the wealth of genome data now available to answer important questions about health and disease.
The Wellcome Trust and Its Founder
The Wellcome Trust is the most diverse biomedical research charity in the world, spending about £450 million every year both in the UK and internationally to support and promote research that will improve the health of humans and animals. The Trust was established under the will of Sir Henry Wellcome, and is funded from a private endowment, which is managed with long-term stability and growth in mind.


