Discrete Distributional Differential Expression (D3E)
D3E is a method for identifying differentially expressed genes from single-cell RNA-seq experiments. D3E compares the full distribution between two sample to identify a set of differentially expressed genes.
[caption id="attachment_104721" align="center" width="1925" class="aligncenter"]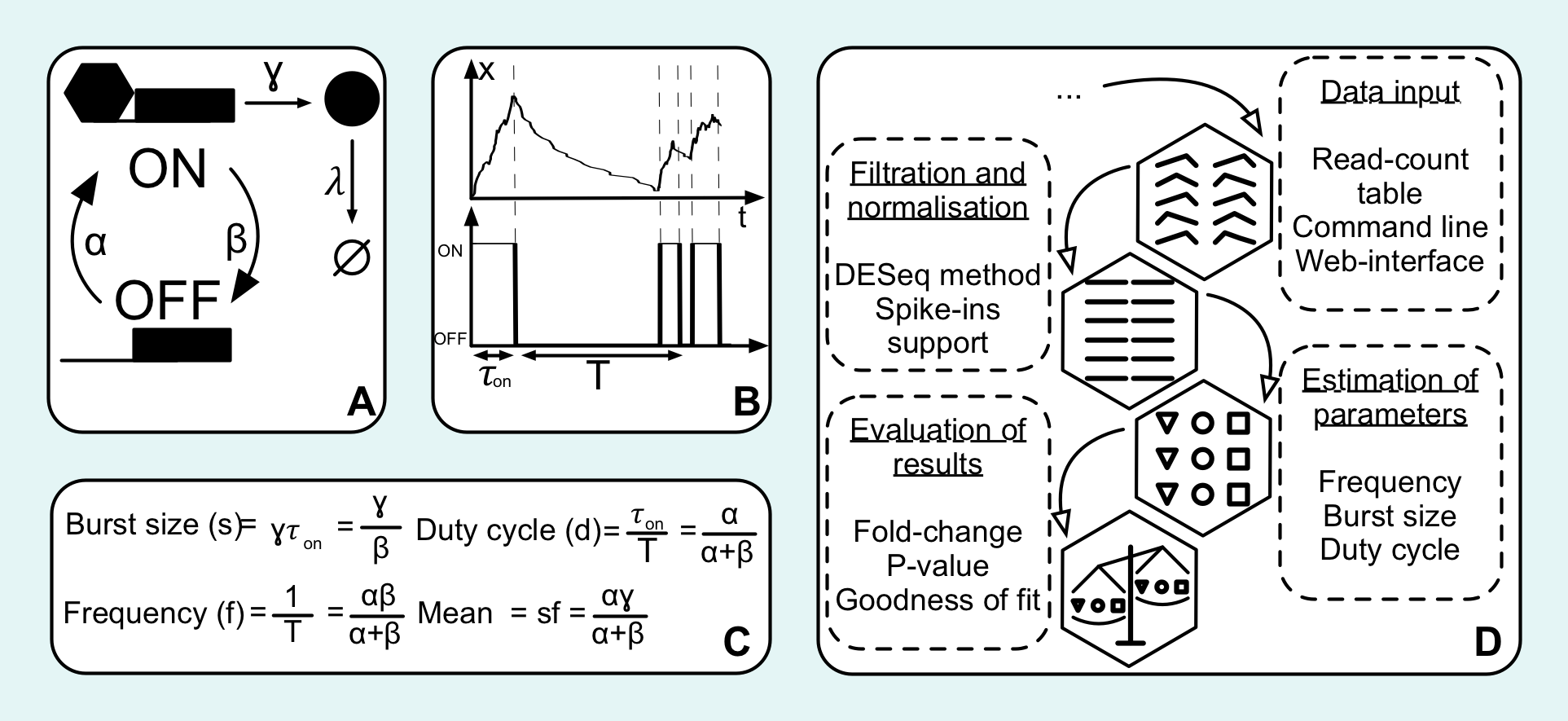 Schematic illustration of the transcriptional bursting model and flowchart of D3E[/caption]We have developed Discrece Distributional Differential Expression (D3E), a method for identifying differentially expressed genes. Obtaining a list of genes which are significantly different between two conditions (e.g. mutant vs wild-type or stimulated vs unstimulated) is central to many transcriptome analyses. D3E is specifically designed for single-cell RNA-seq data, and it identifies differentially expressed genes by comparing the full distribution of expression levels for the two conditions rather than just the mean expression. Furthermore, D3E is based on an analytically tractable stochastic model of gene expression. The model facilitates interpretation of the results, and it makes it possible to generate hypotheses about the underlying mechanisms behind the change in gene expression.
Schematic illustration of the transcriptional bursting model and flowchart of D3E[/caption]We have developed Discrece Distributional Differential Expression (D3E), a method for identifying differentially expressed genes. Obtaining a list of genes which are significantly different between two conditions (e.g. mutant vs wild-type or stimulated vs unstimulated) is central to many transcriptome analyses. D3E is specifically designed for single-cell RNA-seq data, and it identifies differentially expressed genes by comparing the full distribution of expression levels for the two conditions rather than just the mean expression. Furthermore, D3E is based on an analytically tractable stochastic model of gene expression. The model facilitates interpretation of the results, and it makes it possible to generate hypotheses about the underlying mechanisms behind the change in gene expression.
About
Downloads
D3E is available as a command line tool and can be downloaded from github.
Sanger Institute Contributors
Previous contributors

Dr Martin Hemberg
Former CDF Group Leader
