Artemis
Genome browser and annotation tool that allows visualisation of sequence features, next generation data and the results of analyses within the context of the sequence, and also its six-frame translation.
Archive Page
This page is maintained as a historical record and is no longer being updated.
This page was archived in 2022
About
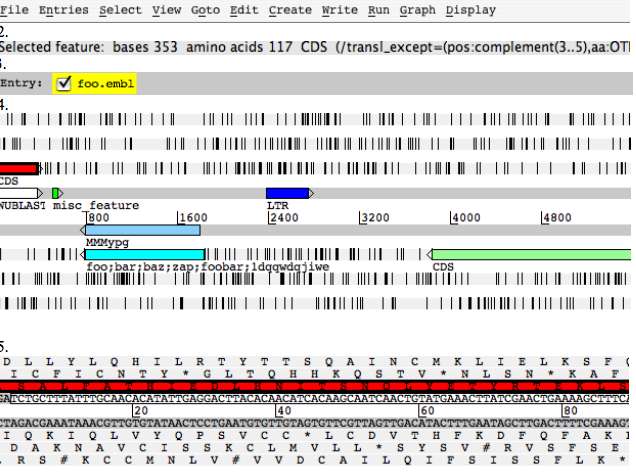
Artemis edit window
Downloads
Please see our GitHub page for download and installation instructions.
Further information
For information and advice on using this software please see the our GitHub page.
In addition, an email discussion list called artemis-users is available and posts to the list since September 2001 are archived at mail-archive.com
Chado
Java Web Start
The Java Web Start facility has now been removed. Please use a locally installed Artemis instead (see Download and Installation section above).
Artemis is available under GPL3.
If you make use of this software in your research, please cite as:
Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data.
Carver T, Harris SR, Berriman M, Parkhill J and McQuillan JA
Bioinformatics (Oxford, England) 2012;28;4;464-9
PUBMED: 22199388; PMC: 3278759; DOI: 10.1093/bioinformatics/btr703
Contact
If you need help or have any queries, please contact us using the details below.
If you need help or have any queries, please contact us using the details below.
- Email: paminfo-help@sanger.ac.uk