The characterisation of somatic mutations allows the possibility of answering a range of questions. I am particularly interested in understanding how different genomic contexts influence mutation and evolution, and how these effects compare at the somatic (either normal or cancer cells) and germline levels. Also of interest is how mutation signatures can help understand cellular processes like DNA repair and how they relate to environmental exposure. The interplay between genomic context and mutational processes is key to unraveling adaptive evolution (causality) during cancer progression. Somatic mutation is also very promising to help understand embryonic development and the organisation of cells within tissues and organs. This is not only important to understanding cancer progression, but also to query the developmental plan for evolutionary adaptive strategies.
My timeline
Joined the Sanger Institute
Staff Scientist at Spanish National Cancer Research Centre
Post-doc Museo Nacional de Ciencias Naturales
I3P post-doctoral fellowship (CNB)
Fundacion BBVA post-doctoral fellowship, Universidad de Vigo & Museo Nacional de Ciencias Naturales
PhD Spanish National Biotecnology Centre (CNB)
MSc Biochemistry and Molecular Biology (UAM)
My publications
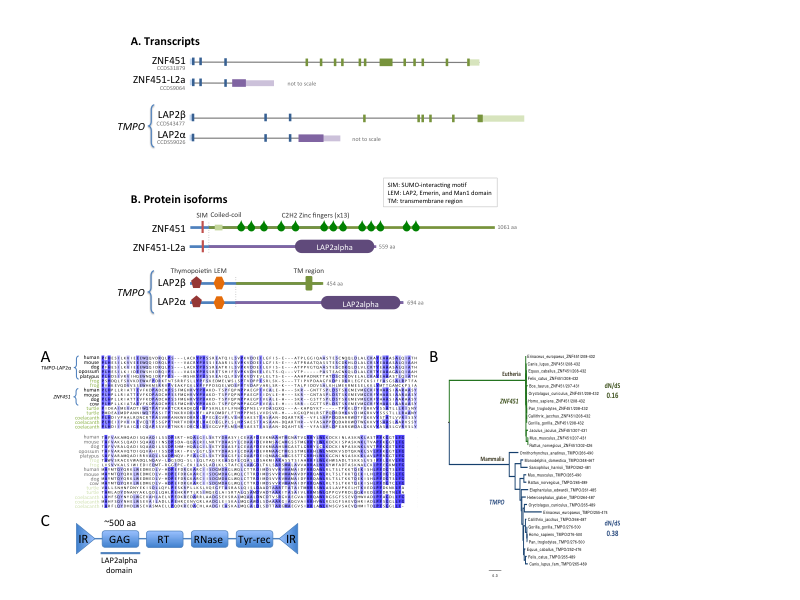
A DIRS1-like retrotransposon inserted within two genes (TMPO/LAP2alpha and ZNF451) in the ancestor of mammals and was co-opted for important new cellular functions. Interestingly, the alternative splicing of the retrotransposed sequence allowed the production of both the new and the untouched original isoforms in the two genes, which may have contributed to the success of this double colonization process.

Proteomics evidence, evolutionary conservation, protein structure and population genetics data strongly suggest that most of the annotated alternative transcripts do not code for functionally relevant proteins
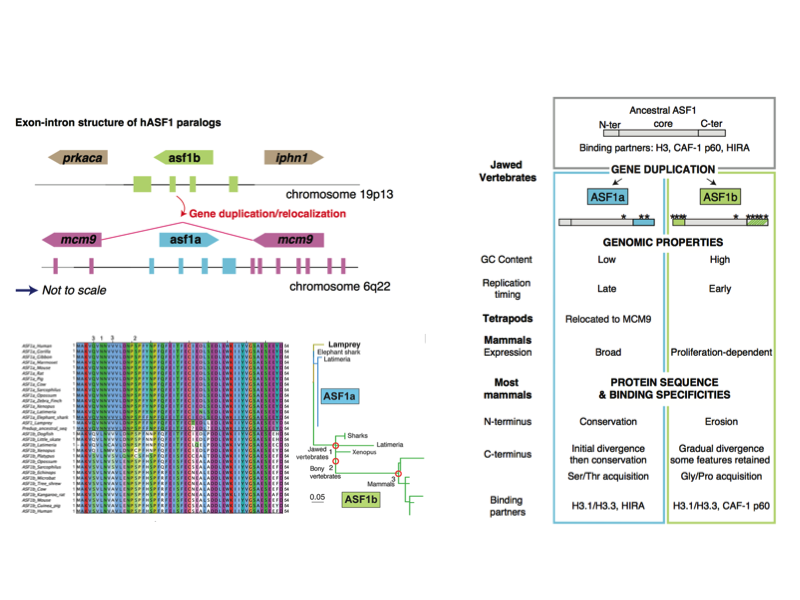
Genes within genes... The subfunctionalization experienced by histone chaperones ASF1a and ASF1b in vertebrates has been influenced by genomic context and has been driven by adaptive evolution. Interestingly, ASF1a relocated into another gene (MCM9) in the ancestor of tetrapods. This relocation possibly had an additional adaptive value as both genes show opposite cell-cycle regulation programs, matching the subfunctionalization of ASF1a.
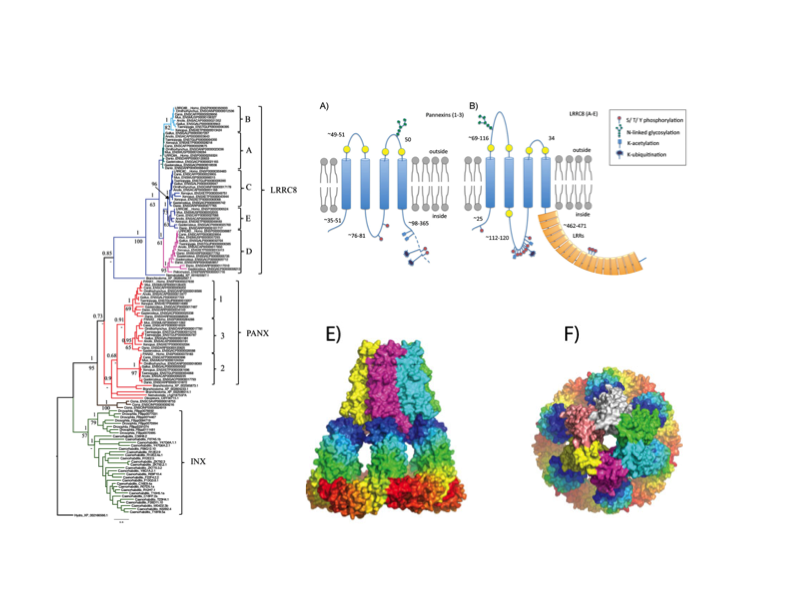
We proposed that LRRC8 proteins share a common ancestor with pannexins and may form hexameric channels involved in cell-cell communication. Recent studies have confirmed the proposed membrane topology and channel activity, revealing that LRRC8 are essential components of volume-regulated anion channels.