Fighting the spread of drug-resistant malaria with genetic surveillance
A large collaborative project of in-depth genomic surveillance of malaria has identified and tracked drug resistant malaria parasites in the Greater Mekong Subregion*, helping inform public health decisions in Southeast Asia.
The project, known as GenRe-Mekong, involves researchers from the Wellcome Sanger Institute, the University of Oxford, MalariaGEN, and several partners in the Greater Mekong Subregion, including multiple National Malaria Control Programmes. It has gathered data from some of the most remote areas of the Greater Mekong Subregion to give a clearer genetic picture of drug resistance in malaria.
In the latest paper, published today (10 August 2021) in eLife, the GenRe-Mekong group describe how they developed and implemented the platform for genetic surveillance of malaria in Southeast Asia, where drug resistance in malaria parasites is an urgent issue. They also describe how the data are used by local public health agencies to plan interventions in malaria endemic areas.
Malaria continues to be a major cause of mortality in many tropical countries, particularly in sub-Saharan Africa, and efforts are ongoing to eliminate the parasite, Plasmodium falciparum, which causes the most severe form of disease. Even though the Greater Mekong Subregion is an area of relatively low P. falciparum malaria prevalence and mortality, drug resistant strains of the parasite have repeatedly arisen from this region and migrated into Asian and African countries, undoing years of progress against the disease, and costing many lives**. Hence, malaria elimination has become an urgent priority in the Greater Mekong Subregion.
In collaboration with the Wellcome Sanger Institute, the University of Oxford, and MalariaGEN, the GenRe-Mekong project developed the SpotMalaria genetic surveillance platform that analyses the collected blood samples and provides a broad range of detailed genetic information about each sample. The project has processed 9,623 blood samples from symptomatic patients across eight countries, the majority of which originated from Vietnam, Laos, Cambodia, Thailand, and Myanmar.
GenRe-Mekong also processed samples from Bangladesh, India, and the Democratic Republic of Congo. SpotMalaria processing only requires small dried blood spot samples, which are easy to collect at public health facilities as part of routine treatment, making it possible to obtain data from the most remote and resource-poor parts of these countries.
The DNA analysis, which was primarily conducted at the Wellcome Sanger Institute, uses high-throughput technologies to extract large amounts of parasite genetic information from each sample. The results are used to create Genetic Report Cards, which are regularly delivered to National Malaria Control Programmes to keep them updated about changes in the parasite population in their country. National Malaria Control Programmes are involved throughout the entire process, from sampling strategy planning, and sample collection, through to joint analyses of results.
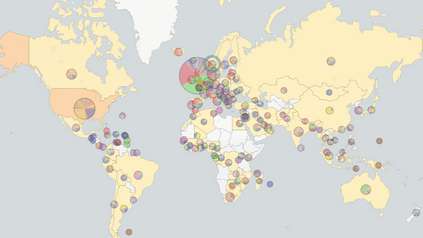
GenRe-Mekong uses the key genetic mutation data extracted to identify areas of parasite resistance to specific drugs, and then collates the results, creating a map showing the prevalence of resistant parasites. These results have helped inform public health agencies, notably influencing decisions in Laos and Vietnam***, such as the choice of frontline therapy, or highlighting which areas require special interventions.
In the future, the project will continue to support National Malaria Control Programmes in their elimination efforts, widening its scope to include analyses of other aspects of malaria parasite epidemiology, such as monitoring outbreak events, or mapping imported strains.
“Drug resistant P. falciparum malaria is an important threat to malaria control, particularly in the Greater Mekong Subregion, where an intense effort for malaria elimination is ongoing, and will hopefully be successful before the parasite becomes close to untreatable. The GenRe-Mekong project works closely with local malaria control programmes, so that they have direct access to the data, facilitating informed decisions on antimalarial drug choices, malaria control and elimination strategies.”
Professor Arjen Dondorp, Deputy Director and Head of Malaria and Critical Care at the Mahidol-Oxford Tropical Medicine Research Unit (MORU) in Bangkok, Thailand
“The technology used in this research was created to make genomic surveillance as accessible as possible, so that information could be gathered from some of the region’s most remote locations. By using a single drop of blood for analysis and developing a technology that has low processing costs, it is possible to undertake these large-scale genetic epidemiology surveys. This method could be used across more countries in different endemic regions to help track malaria cases and inform more public health decisions.”
Dr Christopher Jacob, first author and Product Management Lead at the Wellcome Sanger Institute
“Malaria is still a major killer disease across the world, and it is great to see what an impact genetic surveillance data can have when it is translated into actionable messages that inform public health decisions. This is the first time that such detailed malaria genomic surveillance has been implemented across multiple countries, and it is remarkable how National Malaria Control Programmes and other partners in the Greater Mekong Subregion are working together by sharing data across the region, making this project far more than the sum of its parts.”
Professor Olivo Miotto, Principal Investigator of GenRe-Mekong at Mahidol-Oxford Tropical Medicine Research Unit (MORU) in Bangkok, Thailand, and Associate Professor at the University of Oxford
More information
*This region includes Cambodia, the Lao People’s Democratic Republic, Myanmar, Thailand, Vietnam, and parts of the People’s Republic of China.
** Mita T, Tanabe K, Kita K. (2009) Spread and evolution of Plasmodium falciparum drug resistance. Parasitology International. 58(3): 201-9.
**Trape JF, Pison G, Preziosi MP, et al. (1998) Impact of chloroquine resistance on malaria mortality. C R Acad Sci III 321(8): 689-97.
***Please see the case study sections of the paper for more information on this
Access to the public dataset can be found here https://www.malariagen.net/parasite/genre-mekong#data
Publication:
C Jacob, et al. (2021) Genetic surveillance in the Greater Mekong Subregion and South Asia to support malaria control and elimination. eLife publications. DOI: 10.7554/eLife.62997
Funding:
This research was funded by the Bill & Melinda Gates Foundation, Wellcome, the UK Medical Research Council, UK Department for International Development, and NIAID.