Comparison holds key to human infection
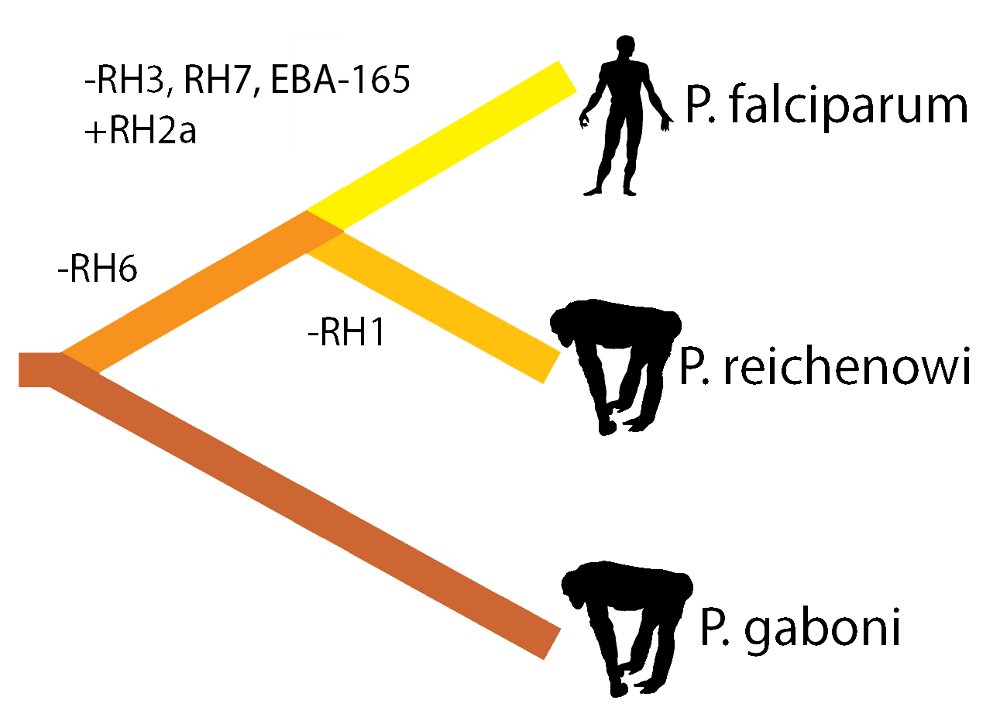
Out of a genome of approximately 5,500 genes, researchers found that most genes have directly equivalent counterparts between the human and primate parasites. However, portions of the P. falciparum genome that differed most profoundly from the P. reichenowi parasite that infects chimpanzees were found to encode proteins that help the parasite to bind to and invade red blood cells, which is where the parasite grows and multiplies.
“Discovering that the key differences lie in genes responsible for red blood cell invasion reassures us that we’ve been looking in the right place. Researchers have identified surface proteins as promising vaccine candidates already; and our finding adds more support, showing that it is the difference in the parasites’ surface proteins that determine which host it will infect.”
Dr Thomas Otto First author at the Wellcome Trust Sanger Institute
This is the first time that an essentially complete genome has been produced for a malaria parasite that infects such a close relative of humans. It provides the first systematic view of the differences between parasites that infect humans and those that infect our close relatives. Human malaria emerged from the Great Apes, so this comparison using chimpanzee malaria is the closest that scientists have come to a full catalogue of the changes associated with parasites switching from our primate relatives into humans.
Plasmodium parasites export proteins to the surface of red blood cells, allowing infected red blood cells to stick to the wall of blood vessels. In human malaria, the best characterised of these proteins are encoded by a highly variable family of genes, allowing the parasites to evade the host immune response and continue the infection. Surprisingly basic rules about this gene family are preserved between chimpanzee and human malaria: despite huge variation in the individual sequence of these surface antigen genes, their absolute numbers and the numbers of sub-types are remarkably preserved. By contrast, other surface antigen repertoires differed very significantly in their numbers.
“Since P. reichenowi and P. falciparum split apart, the major surface antigen gene family has not expanded or contracted; it’s locked at some kind of optimised level.”
Dr Matt Berriman Senior author at the Sanger Institute
DNA used for this research was obtained by the Centre for Disease Control from a chimpanzee infected with a strain of P. reichenowi isolated in the 1950s. This chimpanzee was subsequently cured of the malaria infection. Additional blood samples were collected from orphaned infant chimpanzees infected in the wild with a similar parasite called P. gaboni. The samples were obtained from chimpanzees undergoing routine health checks at a primate sanctuary in Gabon, West Africa.
More information
Funding
This work was supported by the Wellcome Trust (grant number WT 098051) with additional funding to TDO from the European Community’s Seventh Framework Programme (FP7/2007-2013), under grant agreement Number 242095; JR, from the National Institutes of Health (R01 AI091595); BO from CIRMF; FR and FP, from CNRS and IRD; FR, BO, and FP from the Agence Nationale de la Recherche (grant ANR JCJC SVSE 7-2012 ORIGIN); DC from an ERC Advanced Award (grant number 294428); CIN from the Wellcome Trust (grant number WT 082130/Z/07/Z).
Participating Centres
Please see the paper for a full list of participating centres.
Publications:
Selected websites
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


