Antibiotic resistant typhoid detected in countries around the world
“The data was produced by a consortium of 74 collaborators from the leading laboratories working on typhoid and describes one of the most comprehensive sets of genome data on a single human infectious agent. It represents global co-operation in the scientific community at its best. Typhoid affects around 30 million people each year and global surveillance at this scale is critical to address the ever-increasing public health threat caused by multidrug resistant typhoid in many developing countries around the world.”
Dr Vanessa Wong First author from the Wellcome Trust Sanger Institute
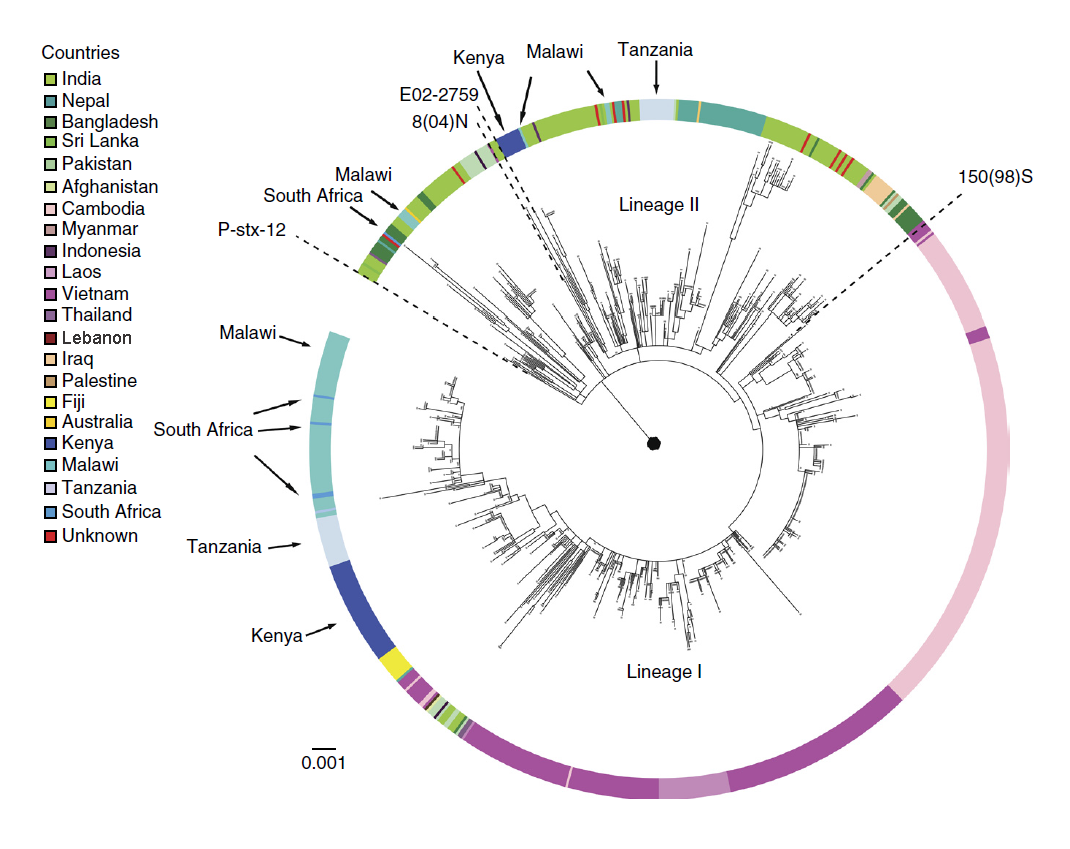
The study shows the H58 clade of Typhi is displacing other typhoid fever strains that have been established over decades and centuries throughout the typhoid endemic world, completely transforming the genetic architecture of the disease. Multidrug resistant H58 has spread across Asia and Africa over the last 30 years, and created a previously underappreciated and ongoing epidemic through countries in eastern and southern Africa with important public health consequences.
Vaccination to prevent the disease is not currently in widespread use in these countries; instead the disease is controlled mainly through use of antimicrobial drugs. H58 Typhi is often resistant to the first-line antimicrobials commonly used to treat the disease, and is continuing to evolve as it spreads to new regions and populations, acquiring novel mutations providing resistance to newer antimicrobial agents, such as ciprofloxacin and azithromycin.
“Multidrug resistant typhoid has been coming and going since the 1970s and is caused by the bacteria picking up novel antimicrobial resistance genes, which are usually lost when we switch to a new drug. In H58, these genes are becoming a stable part of the genome, which means multiply antibiotic resistant typhoid is here to stay.”
Dr Kathryn Holt Senior author from the University of Melbourne
“H58 is an example of an emerging multiple drug resistant pathogen which is rapidly spreading around the world. In this study we have been able to provide a framework for future surveillance of this bacterium, which will enable us to understand how antimicrobial resistance emerges and spreads intercontinentally, with the aim to facilitate prevention and control of typhoid through the use of effective antimicrobials, introduction of vaccines, and water and sanitation programmes.”
Professor Gordon Dougan Senior author from the Sanger Institute
The publication of this research in Nature Genetics coincides with the 9th International Conference on Typhoid and invasive Non-Typhoidal Salmonelloses held by the Coalition against Typhoid (CaT), where these results will be shared with the scientific community. The meeting brings together an international group of healthcare and public health experts, researchers and clinicians to focus on strategies to counteract the spread of typhoid in endemic countries.
“These results reinforce the message that bacteria do not obey international borders and any efforts to contain the spread of antimicrobial resistance must be globally coordinated.”
Dr Stephen Baker An author from The Hospital for Tropical Diseases, an Oxford University Clinical Research Unit in Ho Chi Minh City, Vietnam
More information
Funding
A full list of funding agencies can be found in the publication.
Participating Centres
A full list of participating centres can be found in the publication.
Publications:
Selected websites
Oxford University Clinical Research Unit, Viet Nam
Oxford University Clinical Research Unit in Viet Nam (OUCRU) was established in 1991 and is one of the Wellcome Trust Major Overseas Programmes. It is based within the Ho Chi Minh City (HCMC) Hospital for Tropical Diseases (HTD), at tertiary referral hospital for infectious diseases for southern Viet Nam, under the direction of the Health Service of Ho Chi Minh City and the Ministry of Health. OUCRU also has a base in the capital Hanoi at the National Institute of Infectious & Tropical Diseases (NHTD) and has satellite research units in Kathmandu (Nepal) and Jakarta (Indonesia).
Coalition against Typhoid
The Coalition against Typhoid (CaT) is a global forum of health and immunisation experts working to expedite and sustain rational, evidence-based decisions at the global, regional, national and municipal levels regarding the use of typhoid vaccines to prevent enteric fever. CaT aims to define barriers to the adoption of typhoid vaccines in communities that would benefit most and the key activities that are needed to overcome them.
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


