Genetics underpinning antimalarial drug resistance revealed
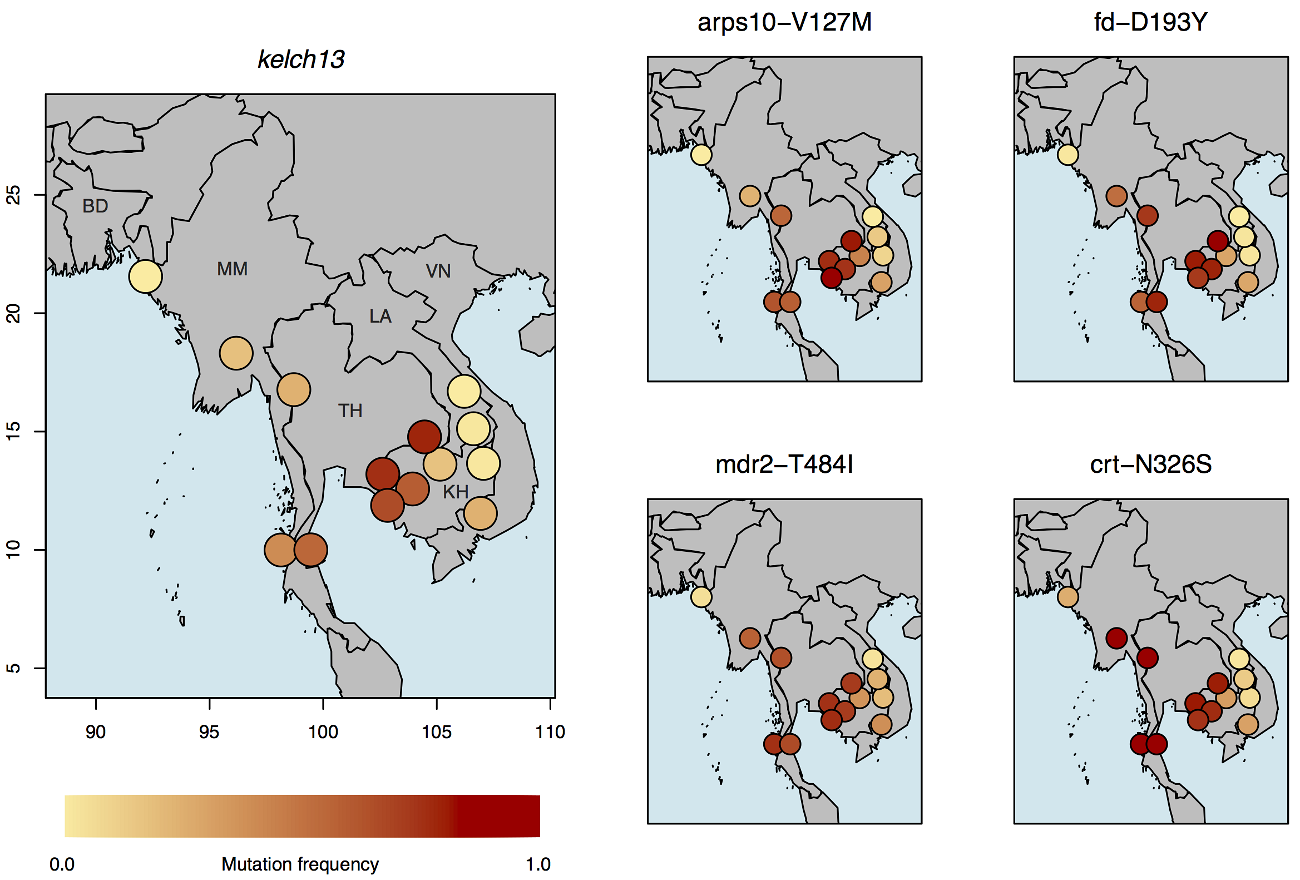
The global research collaboration analysed 1612 samples from 15 locations in Southeast Asia and Africa finding 20 mutations in the kelch13 gene, a known artemisinin resistance marker, that appear to work in concert with a set of background mutations in four other genes to support artemisinin resistance.
“Our findings suggest that these background mutations emerged with limited impact on artemisinin resistance – until mutations occurred in the kelch13 gene. It’s similar to what we see with pre-cancerous cells which accumulate genetic changes but only become malignant when they acquire critical driver mutations that kick-off growth.”
Dr Roberto Amato A first author and Research Associate in Statistical Genomics at the Wellcome Trust Sanger Institute and Oxford University’s Wellcome Trust Centre for Human Genetics
The variety of kelch13 mutations associated with artemisinin resistance, with new variants continually emerging, makes it difficult to use this gene alone as a marker for genetic surveillance.
Monitoring parasite populations for a specific genetic background – in this case, a fixed set of four well-defined mutations in the fd, arps10, mdr2, and crt genes – could allow researchers to assess the likelihood of new resistance-causing mutations emerging in different locations, helping to target high-risk regions even before resistant parasites take hold.
“We are at a pivotal point for malaria control. While malaria deaths have been halved, this progress is at risk if artemisinin ceases to be effective. We need to use every tool at our disposal to protect this drug. Monitoring parasites for background mutations could provide an early warning system to identify areas at risk for artemisinin resistance.”
Nick Day Director of the Mahidol-Oxford Tropical Medicine Research Unit (MORU) in Bangkok, Thailand
Researchers also uncovered new clues about how artemisinin resistance has evolved in Southeast Asia. By comparing parasites from Cambodia, Vietnam, Laos, Thailand, Myanmar and Bangladesh, scientists found that the distribution of different kelch13 mutations are localised within relatively well-defined geographical areas.
Whilst artemisinin resistant parasites do appear to have migrated across national borders, this only happened on a limited scale and, in fact, the most widespread kelch13 mutation, C580Y, appeared to have emerged independently on several occasions. Notably parasites along the Thailand-Myanmar border appear to have acquired this mutation separately from those in Cambodia and Vietnam. Crucially, parasite populations in both regions possess the genetic background mutations, even though they are clearly genetically distinct.
There remain many unanswered questions.
“We don’t yet know the role of these background mutations. Some may not affect drug resistance directly, but rather provide an environment where drug resistance mutations are tolerated. Since kelch13 has hardly changed in 50 million years of Plasmodium evolution, we can assume that this gene is essential to parasite survival. Therefore, kelch13 mutations may severely handicap mutant parasites, compromising their survival unless some other change can counteract this negative effect.”
Dr Olivo Miotto A first author and Senior Informatics Fellow at MORU and the Centre for Genomics and Global Health
Mutations in the kelch13 gene were present, yet rare, in Africa but weren’t associated with artemisinin resistance and lacked the genetic background present in artemisinin-resistant parasites in Southeast Asia. This provides some reassurance for public health authorities working to prevent the spread of artemisinin resistance to Africa where most malaria deaths occur.
“These data serve as a reminder of how crucial surveillance and elimination programmes are. At present artemisinin resistance appears to be largely confined to Southeast Asia but the situation might change as the parasite population continues to evolve. By linking genomic data with clinical data we’re developing a better understanding of the multiple genetic factors involved in the emergence of resistance, and that is starting to provide vital clues about how to prevent its spread.”
Professor Dominic Kwiatkowski Head of the Malaria Programme at the Wellcome Trust Sanger Institute and Professor of Genomics and Global Health at Oxford University
More information
Funding
The sequencing for this study was funded by the Wellcome Trust through core funding of the Wellcome Trust Sanger Institute (098051). The Wellcome Trust also supports the Wellcome Trust Centre for Human Genetics (090532/Z/09/Z), the Resource Centre for Genomic Epidemiology of Malaria (090770/X/09/Z), and the Wellcome Trust Mahidol University Oxford Tropical Medicine Research Programme (0892375/H/09/Z). The Centre for Genomics and Global Health is supported by the Medical Research Council (G0600718). This work was funded in part by the Bill & Melinda Gates Foundation (OPP1040463), the Intramural Research Program of the NIAID, NIH, and the UK Department for International Development (PO5408).
Participating Centres
A full list of participating centres can be found on the paper.
Publications:
Selected websites
MalariaGEN
The Malaria Genomic Epidemiology Network (MalariaGEN) is a data-sharing community working to develop new tools to control malaria by integrating epidemiology with genome science.
Wellcome Trust Centre for Human Genomics
The Wellcome Trust Centre for Human Genomics is a research institute of the Nuffield Department of Medicine at the University of Oxford, funded by the University, the Wellcome Trust and numerous other sponsors. With more than 400 active researchers and around 70 employed in administrative and support roles, the Centre is an international leader in genetics, genomics and structural biology. We collaborate with research teams across the world on a number of large-scale studies in these areas. Our researchers expend close to £20m annually in competitively-won grants, and publish around 300 primary papers per year.
MRC Centre for Genomics and Global Health
Through a number of international collaborations, the MRC Centre for Genomics and Global Health is integrating genomic and population genetic data with clinical and epidemiological data in order to develop innovative methods to monitor evolutionary changes in the pathogens that cause disease and the vectors that transmit them. This type of genomic surveillance can provide useful reconnaissance for disease control efforts while also generating discoveries of fundamental scientific interest.
Mahidol Oxford Tropical Medicine Research Unit
The Mahidol Oxford Tropical Medicine Research Unit (MORU) develops effective and practical means of diagnosing and treating malaria and other neglected diseases such as melioidosis, typhus, TB and leptospirosis. MORU carries out targeted clinical and public health research that provides appropriate and affordable interventions that result in measurable improvements in the health of people living in the resource-poor tropics. A research collaboration between Mahidol University (Thailand), Oxford University (UK) and the UK’s Wellcome Trust, MORU has study sites and collaborations across seven countries in Asia and Africa.
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


