Project finds 12 new genetic causes of developmental disorders
DDD, which is a collaboration between the NHS and the Wellcome Trust Sanger Institute and is funded by the Department of Health and Wellcome Trust through the Health Innovation Challenge Fund, worked with 180 clinicians from 24 regional genetics services across the UK and the Republic of Ireland to analyse all ~20,000 genes in each of 1133 children with severe disorders so rare and poorly characterised that they cannot be easily diagnosed using standard clinical tests. The benefits of diagnosis include improving clinical management, helping parents obtain support, informing reproductive choice and providing a molecular basis for the disorder, which is the starting point in the search for new treatments.
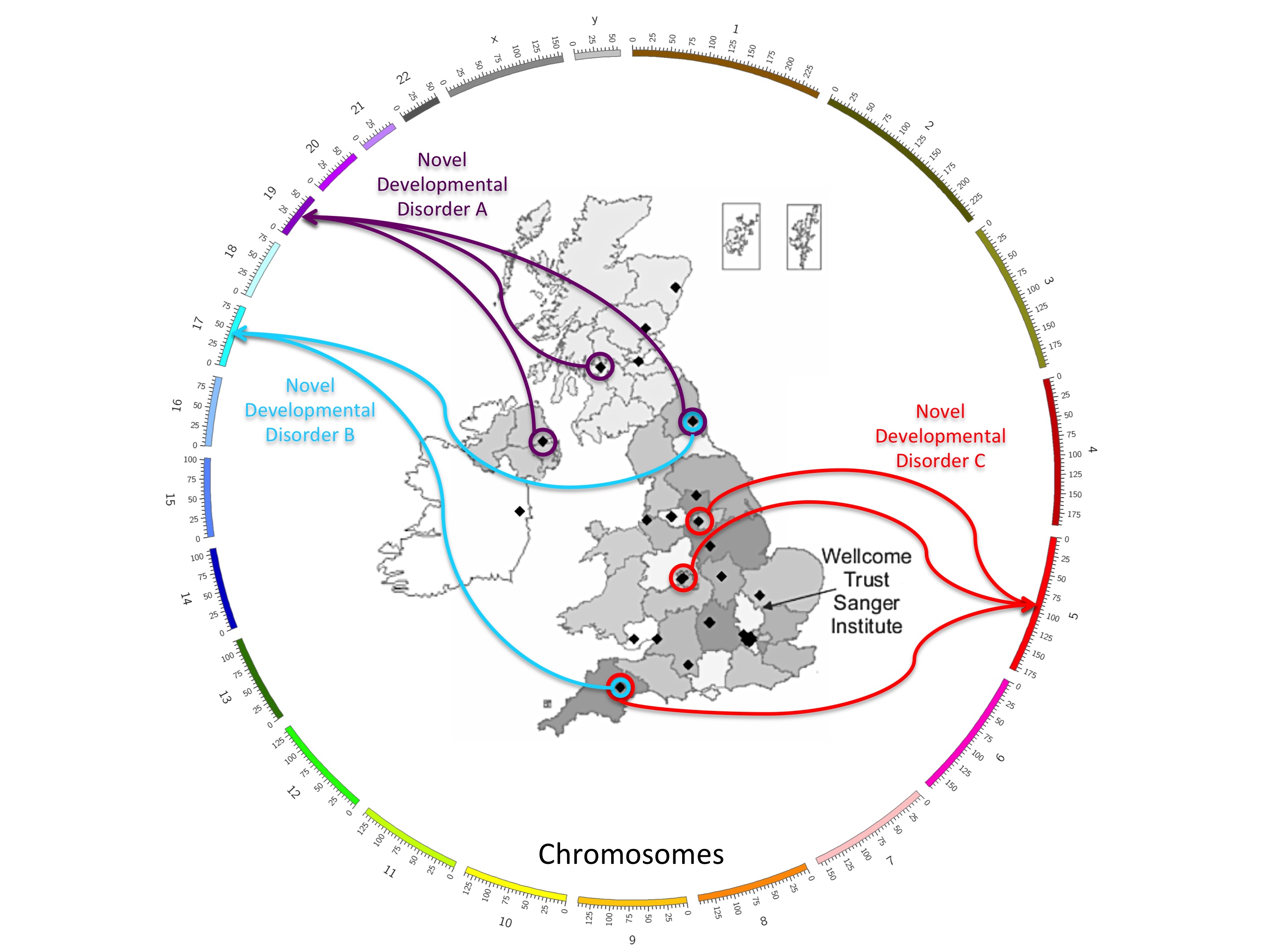
The DDD project works by collecting together clinical information in a database along with the genetic variants from each patient’s genome. If patients who share similar symptoms also have variants in common, it helps to narrow down the search for causative mutations across the genome. However, this can be challenging, since the chance of having a particular type of mutation can be as low as one in fifty million. DDD’s nationwide secure data-sharing network has made it possible to find and compare these incredibly rare disorders; in fact, for four of the 12 newly identified genes, identical mutations were found in two or more unrelated children living hundreds of miles apart.
“Working at enormous scale, both nationwide and genome-wide, is critical in our mission to find diagnoses for these families. This project would not have been possible without the nationwide reach of the UK National Health Service, which has enabled us to unite a number of families who live hundreds of miles apart but whose children share equivalent mutations and very similar symptoms.”
Dr Helen Firth An author from the Department of Clinical Genetics at Addenbrooke’s Hospital and Clinical Lead for the DDD study
In one example, two unrelated children, both with identical mutations in the gene PCGF2, which is involved in regulating genes important in embryo development, were found to have strikingly similar symptoms and facial features. This constitutes the discovery of a new, distinct dysmorphic syndrome.
All of the newly discovered developmental disorders were caused by new, ‘de novo‘, mutations, which are present in the child but are not in their parents’ genomes. The DDD project has shown that it is critical to use, where possible, genetic data from parents, most of whom do not have a developmental disorder, to help filter out benign inherited variants and find the cause of their child’s condition.
“The DDD study has shown how combining genetic sequencing with more traditional strategies for studying patients with very similar symptoms can enable large-scale gene discovery. This data-set becomes more effective with each diagnosis and each newly identified gene.”
Professor Sir John Burn Professor of Clinical Genetics at Newcastle University
Originally, the DDD project focused on applying array technology to screen genes for deletions or duplications that cause the patient’s disorder. However, this strategy enabled researchers to find a diagnosis for only 5 per cent of patients. Improvements in sequencing technology have allowed DDD to use genome-wide ‘exome’ sequencing that searches through all protein-coding genes for all classes of genetic variants. This approach produces 100 times more data but delivers a diagnosis for 30 per cent of patients.
“The success of DDD has provided a valuable test bed for Genomics England. This research has shown that the Government’s commitment to sequencing 100,000 genomes can produce powerful data that will make a real difference to genetic research as well as to clinical diagnostics and treatment.”
Professor Mark Caulfield Chief Scientist for Genomics England Limited
Nonetheless, some DDD children will not be able to be diagnosed by looking at data from UK patients in isolation, and so to identify similar patients from around the world DDD is sharing limited anonymised genetic and clinical data on these undiagnosed DDD children through the DECIPHER database (https://www.deciphergenomics.org). Researchers hope that the project will inspire more clinical and research programmes around the world to deposit data in the DECIPHER database to pinpoint more genetic causes of developmental disorders and improve diagnostic rates internationally.
“There is a clear moral imperative for both clinical testing laboratories and research studies to share this information globally. DDD and DECIPHER have demonstrated that large-scale data sharing can give families the diagnoses they so urgently need; diagnoses that simply cannot be made by looking at the data in isolation.”
Dr Matt Hurles Senior author and principal investigator on the DDD project from the Sanger Institute
More information
Funding
Generation Scotland has received core funding from the Chief Scientist Office of the Scottish Government Health Directorates CZD/16/6 and the Scottish Funding Council HR03006. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund [grant number HICF-1009-003], a parallel funding partnership between the Wellcome Trust and the Department of Health, and the Wellcome Trust Sanger Institute [grant number WT098051]. The research team acknowledges the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network.
Participating Centres:
A full list of participating centres can be found on the paper.
Publications:
Selected websites
The Health Innovation Challenge Fund
The Health Innovation Challenge Fund is a parallel funding partnership between the Wellcome Trust and the Department of Health to stimulate the creation of innovative healthcare products, technologies and interventions and to facilitate their development for the benefit of patients in the NHS and beyond.
The Department of Health (DH)
The Department of Health (DH) helps people to live better for longer. The Department leads, shapes and funds health and care in England, making sure people have the support, care and treatment they need, with the compassion, respect and dignity they deserve. The Department funds health research and encourages the use of new technologies because it’s important to the development of new, more effective treatments for NHS patients. Innovation is needed so that decisions about health and care are based on the best and latest evidence.
The National Institute for Health Research (NIHR)
The National Institute for Health Research (NIHR) is funded by the Department of Health to improve the health and wealth of the nation through research. Since its establishment in April 2006, the NIHR has transformed research in the NHS. It has increased the volume of applied health research for the benefit of patients and the public, driven faster translation of basic science discoveries into tangible benefits for patients and the economy, and developed and supported the people who conduct and contribute to applied health research. The NIHR plays a key role in the Governments’ strategy for economic growth, attracting investment by the life-sciences industries through its world-class infrastructure for health research. Together, the NIHR people, programmes, centres of excellence and systems represent the most integrated health research system in the world.
Deciphering Developmental Disorders (DDD UK)
The aim of the Deciphering Developmental Disorders study is to advance clinical genetic practice for children with developmental disorders by the systematic application of the latest microarray and sequencing methods while addressing the new ethical challenges raised.
Newcastle University
Genomics England Limited
Genomics England, with the consent of participants and the support of the public, is creating a lasting legacy for patients, the NHS and the UK economy through the sequencing of 100,000 genomes: the 100,000 Genomes Project. Genomics England was set up by the Department of Health to deliver the 100,000 Genomes Project. Initially the focus will be on rare disease, cancer and infectious disease. The main phase of the programme will begin in 2015 and will be completed by the end of 2017.
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


