Finding the genetic culprits that drive antibiotic resistance
Researchers have developed a powerful new tool to identify genetic changes in disease-causing bacteria that are responsible for antibiotic resistance. The results from this technique could be used in clinics within the next decade to decide on the most effective treatments for diseases such as pneumonia and meningitis.
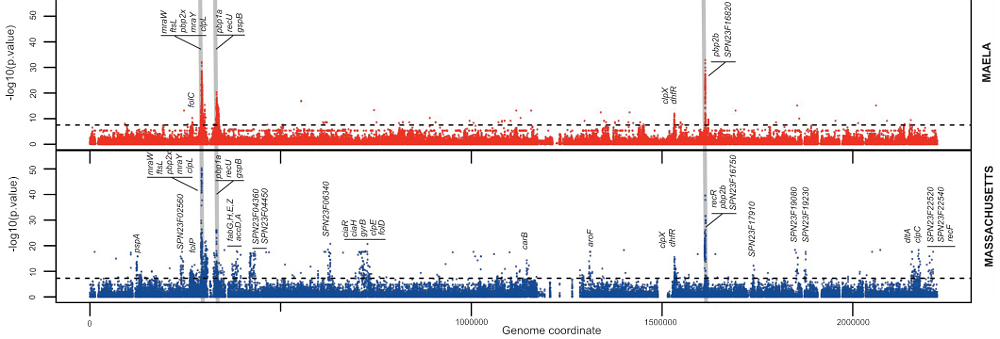
The team looked at the genome of Streptococcus pneumoniae, a bacterial species that causes 1.6 million deaths worldwide each year. In the most detailed research of its kind, scientists used a genome-wide association study (GWAS) to locate single-letter changes in the DNA code of the bacterium, which enable it to evade antibiotic treatment.
While GWAS has been used for a decade to identify gene function in humans, it was thought to be impossible to use the technique on bacterial DNA until now.
“The results of this research are very interesting. For the first time, we are able to see, at large scale, causative variants that allow bacteria such as Streptococcus pneumoniae to resist our efforts to treat and control it.
“We can begin to see how this might help us to develop more effective treatment strategies in the near future.”
Claire Chewapreecha First author from the Wellcome Trust Sanger Institute
GWAS studies search through the genome for locations where single DNA changes are associated with properties of the organism, like antibiotic resistance. For this to work, it is essential that a genetic exchange process called recombination has occurred. This is where two DNA sequences combine, exchanging genetic data, and shuffling combinations of single changes. Because recombination, which is very common in humans, is rare in bacteria, researchers have previously been unable to locate the individual changes in the sequence of bases that make up a gene. Until now it has only been possible to locate the general area where change has occurred in so-called mosaic genes comprised of genetic data from multiple strains of bacteria.
To overcome this hurdle, researchers used the richest available set of data for S. pneumoniae, which was collected by Dr Claudia Turner, a Research Paediatrician, and Dr Paul Turner, a Clinical Microbiologist, at the Mahidol Oxford Tropical Medicine Research Unit. The dataset of over 3,000 samples of Streptococcus pneumoniae isolated from almost 1,000 infants and mothers at a refugee camp on the border between Myanmar and Thailand provided enough recombination events to give researchers precise data about the locations of changes that cause resistance. A second set of several hundred isolates was collected from Massachusetts as part of a project to assess the impact of the vaccine against S. pneumoniae introduced in the USA in 2001.
“In this study we’ve shown that this powerful genetic tool, which has transformed our understanding of human genetics, can be applied to bacteria. This opens up new avenues of research into antibiotic resistance, transmission and virulence that were previously thought impossible in bacterial genomics.”
Professor Stephen Bentley A senior author from the Sanger Institute
The next phase of research will involve fine-tuning the technique to be able to identify genes in bacteria that make strains more virulent and genes that enable transmission of a bacterial strain between hosts. As genetic sequencing moves into clinics, this detailed understanding will inform control and treatment strategies.
“Uncovering all the single-letter differences underlying resistance will be essential for future use of genome sequencing to predict antibiotic sensitivity in clinical microbiology. Genome-wide association studies will enable this by allowing us to pinpoint the location of the real genetic culprits rather than our current mapping that gets us only to the right general area.”
Professor Julian Parkhill Sanger Institute
More information
Funding
Please see the paper for details about funding.
Participating Centres
Please see the paper for a full list of participating centres.
Selected websites
The Shoklo Malaria Research Unit
The Shoklo Malaria Research Unit was established in 1986 in Shoklo refugee camp on the Thai Burma border. It is a field station of the faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand, and is part of the Mahidol-Oxford Research Unit (MORU) supported by the Wellcome Trust (UK).
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


