Library that can determine resistance
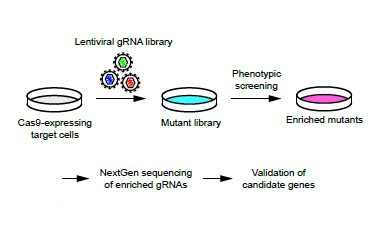
CRISPR technology uses the DNA-cutting enzyme Cas9, with the help of a guide RNA sequence, to find and modify genetic targets. Scientists can easily engineer multiple new guide RNAs using standard molecular biology techniques. This makes for a much faster and efficient method to modify the genome of any cell type in any species.
The team found that a remarkable 50 of 52 guide RNAs tested successfully cut both copies of the genes. The extremely high success rate for these engineered guide RNAs seems to be consistent across many cell types, which led them to create a library of guide RNAs targeting every gene in the mouse genome.
“CRISPR technology is revolutionising how we study the behaviour of cells. We’ve developed a thorough library that can be used by other researchers to study the role of any gene. We can create a library of this type for any cell or any species.”
Dr Kosuke Yusa Lead author from the Wellcome Trust Sanger Institute
The team has engineered a library of nearly 90,000 of these guide RNAs that can be used to target and modify every gene in the mouse genome, and created mutant stem cell libraries. They subjected the mutant libraries to a genetic screen against a bacterial toxin, Clostridium septicum alpha-toxin, to better understand how resistance can occur: This agent has toxic effects on every cell type.
The team targeted 26 genes that are known to be associated with the synthesis of the receptors of this bacterial toxin. They revealed that 17 of these genes were responsible for resistance. Importantly, they also identified previously unknown genes whose mutations confer with resistance to this toxic agent.
“Dr Yusa and his team have used the CRISPR technique in a very innovative and creative way which has allowed them to identify resistance genes for toxins and other agents. This study highlights the power of the technology and illustrates some potential applications it can have.”
Professor Allan Bradley Director Emeritus of the Wellcome Trust Sanger Institute
The team will now use this library to create mutations in cancer cell lines. A major problem with cancer drugs is that often cells can quickly acquire resistance and reject the treatment. By screening for genes that have lost all function through mutation, the team can determine what genes are involved in acquiring resistance to cancer treatments and thus find a clinical target.
More information
Funding
This work was supported by the Wellcome Trust.
Participating Centres
- Wellcome Trust Sanger Institute, Hinxton, Cambridge, CB10 1SA, UK
Publications:
Selected websites
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


