New technique identifies novel class of cancer's drivers
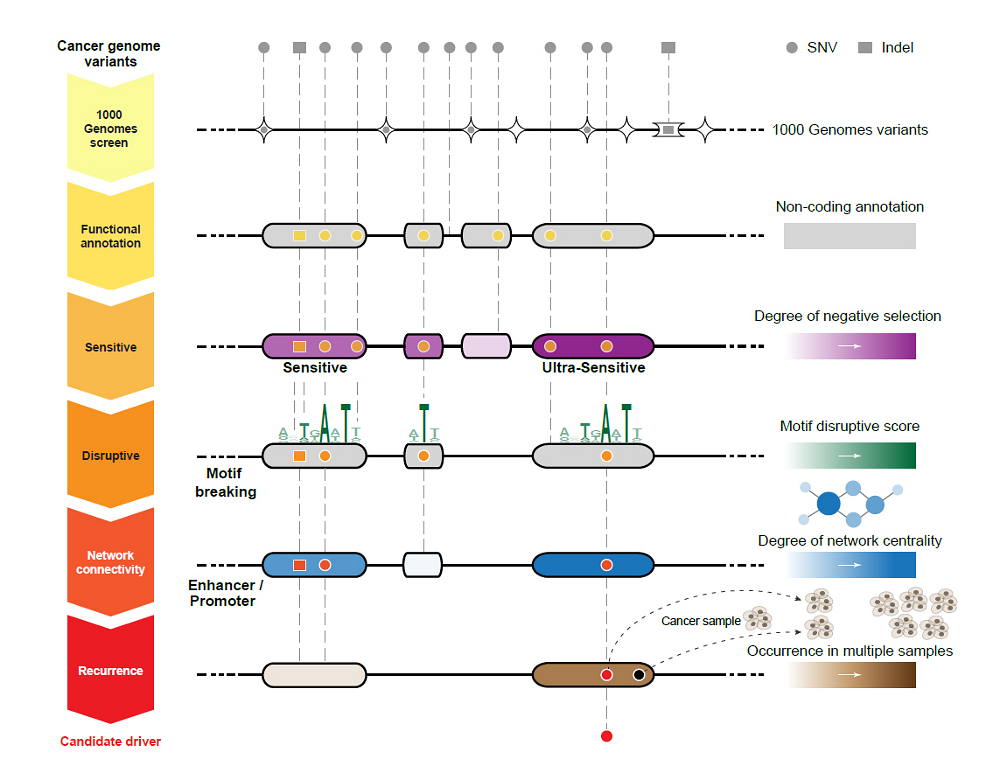
Their approach reveals many potential genetic variants within non-coding DNA that drive the development of a variety of different cancers. This approach has great potential to find other disease-causing variants.
Unlike the coding region of the genome where our 23,000 protein-coding genes lie, the non-coding region – which makes up 98 per cent of our genome – is poorly understood. Recent studies have emphasised the biological value of the non-coding regions, previously considered ‘junk’ DNA, in the regulation of proteins. This new information provides a starting point for researchers to sieve through the non-coding regions and identify the most functionally important regions.
“Our technique allows scientists to focus in on the most functionally important parts of the non-coding regions of the genome. This is not just beneficial for cancer research, but can be extended to other genetic diseases too.”
Professor Mark Gerstein Senior author from the University of Yale
The team used the full set of genetic variants from the first phase of the 1000 Genomes Project, together with information about the non-coding regions generated by the ENCODE Project, and identified regions that did not accumulate much variation.
Protein-coding genes play a crucial role in human survival and fitness, and are under strong ‘purifying’ selection, which removes variation. The team found that some non-coding DNA regions showed almost the same low levels of variation as protein-coding genes, and called these ‘ultrasensitive’ regions.
Within the ultrasensitive regions, they looked at specific single DNA letters that, when altered, caused the greatest disturbance to the genetic region. If this non-coding, ultrasensitive region is central to a network of many related genes, variation can cause a greater knock-on effect, resulting in disease.
They integrated all this information to develop a computer workflow known as FunSeq. This system prioritises genetic variants in the non-coding regions based on their predicted impact on human disease.
“Our method is a practical and successful way to screen for purifying selection in non-coding regions of the genome using freely available data such as those from the ENCODE and 1000 Genomes Projects. It really shows the value of these large-scale open access data-sets.”
Dr Yali Xue Author from the Wellcome Trust Sanger Institute
The team applied FunSeq to 90 cancer genomes including breast cancer, prostate cancer and brain tumours, and found nearly 100 potential non-coding cancer driving variants. In the breast cancer genomes, for example, they found a single DNA letter change that seems to have great impact on the development of breast cancer. This single letter change occurs in an ultrasensitive region that is central to a network of many related genes.
“Although we see that the first effective use of our tool is for cancer genomes, this method can be applied to find any potential disease-causing variant in the non-coding regions of the genome. We are excited about the vast potential of this method to find further disease-causing, and also beneficial variants, in these crucial but unexplored areas of our genome.”
Dr Chris Tyler-Smith Lead author from the Wellcome Trust Sanger Institute
More information
Funding
A full list of funding can be found on the paper
Participating Centres
A full list of participating centres can be found on the paper
Publications:
Selected websites
1000 Genomes Project
The 1000 Genomes Project is the first project to sequence the genomes of a large number of people, to provide a comprehensive resource on human genetic variation.
ENCODE
ENCODE, the Encyclopedia of DNA Elements, is a project funded by the National Human Genome Research Institute to identify all regions of transcription, transcription factor association, chromatin structure and histone modification in the human genome sequence.
GENCODE
The aim of GENCODE as a sub-project of the ENCODE scale-up project is to annotate all evidence-based gene features in the entire human genome at a high accuracy. The result will be a set of annotations including all protein-coding loci with alternatively transcribed variants, non-coding loci with transcript evidence, and pseudogenes.
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


