Vaccine reduces pneumococcal disease but not bacterial population
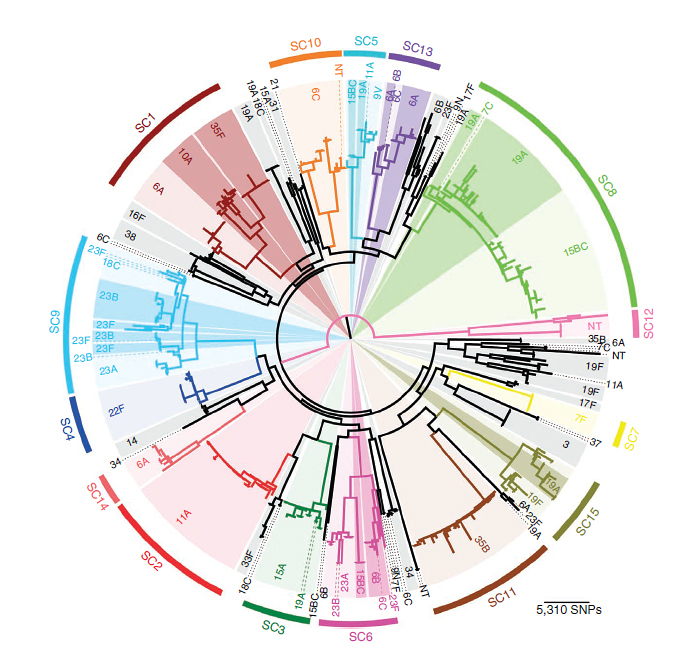
The collaboration between the Wellcome Trust Sanger Institute and the Harvard School of Public Health sequenced the whole genomes of Streptococcus pneumoniae taken from clinical samples in the seven years after the introduction of a pneumococcus vaccine in the US in 2000. The findings reveal that introducing a bacterial vaccine produces a markedly different effect to viral vaccines. In contrast to flu, where the virus evolves to overcome vaccination, other pre-existing strains from the pneumococcus bacterial population fill the void produced by the vaccine. These strains are thought to be less invasive, explaining why disease incidence reduced, but bacterial population numbers and genetic diversity barely altered.
Pneumococcus colonises the human nasal passages where it normally has no detrimental effects. However, for reasons that are not fully understood, the bacteria can cause diseases such as pneumonia, bacteraemia and meningitis. In 2000, 14.5 million episodes of serious pneumococcal disease were estimated to occur, including about 826,000 deaths in children aged 0-5 years (World Health Organization). Since 2000, anti-pneumococcal vaccines have been introduced which have been successful in protecting against seven strains that are most associated with disease in the US.
“This study gives an unprecedented insight into the bacteria living and transmitting among us. We can characterize these bugs to an almost unimaginable degree of detail, and in so doing understand better what helps them survive even in the presence of an effective vaccine.”
Co-senior author William Hanage Associate Professor of Epidemiology at Harvard School of Public Health
The team sequenced the genomes of more than 600 pneumococcal carriage isolates collected in Massachusetts, US through the roll-out of the 7-valent pneumococcal vaccine. The results confirmed that the parts of the bacterial population targeted by the vaccine had almost disappeared, but were replaced by pre-existing, rarer strains. The genetic composition of the new population is very similar to the original one, except for a few genes that were directly affected by the vaccine. This small genetic alteration may be responsible for the large reduction disease rates.
“The widespread use of whole-genome sequencing will allow better surveillance of bacterial populations – even those that are genetically diverse – and improve understanding of their evolution. In this study, we were even able to see how quickly these bacteria transmit between different regions within Massachusetts and identify genes associated with bacteria in children of different ages.”
Marc Lipsitch Professor of Epidemiology at Harvard School of Public Health and co-senior author of the study
The team suggests that further focused sequencing programmes may prove crucial to the future control of this, and other, bacterial pathogens that use similar mechanisms to outsmart human control measures.
“In the near future, we will be able to monitor pathogen evolution in close to real-time. Just as we know that bacteria can acquire antibiotic resistance, we knew that pneumococcus could potentially evade the vaccine. This is a demonstration of how we can monitor the evolution of pneumococcus and the mechanisms at play, information which will be beneficial for vaccine strategists. If we can quickly and precisely trace the emergence of disease-causing bacteria, we may be able to better target interventions to limit the burden of disease.”
Professor Stephen Bentley Co-senior author from the Wellcome Trust Sanger Institute
More information
Funding
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the US National Institutes of Health (NIH) and by the Wellcome Trust.
Participating Centres
- Center for Communicable Disease Dynamics, Department of Epidemiology, Harvard School of Public Health, Boston, Massachusetts, USA.
- Pathogen Genomics, Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK.
- Department of Population Medicine, Harvard Medical School and Harvard Pilgrim Health Care Institute, Boston, Massachusetts, USA.
- Division of General Pediatrics, Boston Children’s Hospital, Boston, Massachusetts, USA.
- Maxwell Finland Laboratory for Infectious Diseases, Boston University Medical Center, Boston, Massachusetts, USA.
- Department of Laboratory Medicine, Boston Children’s Hospital, Boston, Massachusetts, USA.
- Division of Infectious Diseases, Department of Medicine, Boston Children’s Hospital, Boston, Massachusetts, USA.
- Department of Medicine, University of Cambridge, Addenbrooke’s Hospital, Cambridge, UK.
- Department of Immunology and Infectious Diseases, Harvard School of Public Health, Boston, Massachusetts, USA.
Publications:
Selected websites
Harvard School of Public Health
Harvard School of Public Health brings together dedicated experts from many disciplines to educate new generations of global health leaders and produce powerful ideas that improve the lives and health of people everywhere. As a community of leading scientists, educators, and students, we work together to take innovative ideas from the laboratory and the classroom to people’s lives-not only making scientific breakthroughs, but also working to change individual behaviors, public policies, and health care practices. Each year, more than 400 faculty members at HSPH teach 1,000-plus full-time students from around the world and train thousands more through online and executive education courses. Founded in 1913 as the Harvard-MIT School of Health Officers, the School is recognized as America’s first professional training program in public health.
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.