Evolving genes lead to evolving genes
Researchers have designed a method that can universally test for evolutionary adaption, or positive (Darwinian) selection, in any chosen set of genes, using re-sequencing data such as that generated by the 1000 Genomes Project. The method identifies gene sets that show evidence for positive selection in comparison with matched controls, and thus highlights genes for further functional studies.
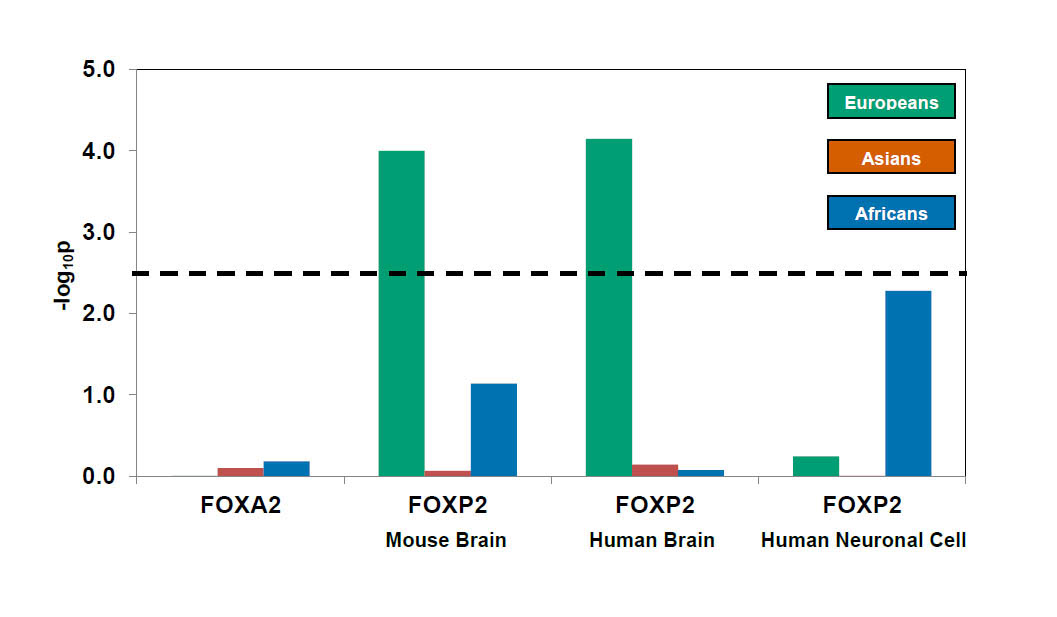
The method was employed to test whether any of the genes directly regulated by FOXP2 may themselves have undergone positive selection following the known selection at the FOXP2 genetic region. Human FOXP2 defects have been implicated in speech and language disorders, and altered versions of the gene have been selected several times during human evolution. Have these evolutionary changes in FOXP2 function or expression exposed its target genes to novel selective pressures?
The study used three sets of genes regulated by FOXP2 that had been identified by previous genomic screens in mice and humans. These sets were compared with matched controls using this method.
“Our method worked well and overall, there was strong evidence for selection of FOXP2-regulated genes in the Europeans, but not in the Asian, or African populations. The subset of FOXP2-regulated genes that were selected in Europeans play roles in neural cell development, cellular signalling, reproduction and immunity.”
Dr Qasim Ayub First author from the Wellcome Trust Sanger Institute
The selection in the Europeans might be due to local adaptations to environment or pathogens. Some of the genes, such as CNTNAP2 and RBFOX1, showed strong signals of selection in all populations examined. Intriguingly, both these genes are highly expressed in the brain and have been implicated in neurodevelopmental disorders, including autism.
“Our study highlights how genes can acquire and adapt to different roles in human evolution. We should never underestimate how complex human biology can be. A next step could be to test whether variants in the selected genes are associated with risk of human neurodevelopmental problems, like language impairments and autism spectrum disorders. Genetic networks can give us powerful insights into the biology underlying these important disorders, which make a major impact on modern human society.”
Professor Simon Fisher A co-author from the Max Planck Institute for Psycholinguistics
“We have already started using this method to look for selection in various other gene sets such as those associated with diabetes and viral infections. Our method is opening new doors to understanding how modern humans have genetically adapted to their local environments and finding candidate genes to study biological function. This approach is a practical and successful way to screen for positive selection and adaptation signals in different gene sets and populations using whole-genome sequencing data.”
Dr Chris Tyler-Smith Lead author from the Wellcome Trust Sanger Institute
More information
Funding
This work was supported by The Wellcome Trust and the Max Planck Society.
Participating Centres
- The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambs. CB10 1SA, United Kingdom
- Language and Genetics Department, Max Planck Institute for Psycholinguistics, Wundtlaan 1, 6525 XD, Nijmegen, The Netherlands
- Donders Institute for Brain, Cognition and Behaviour, Radboud University, 6525 EN, Nijmegen, The Netherlands
- Division of Biological Anthropology, University of Cambridge, Cambridge, CB2 1QH, United Kingdom
Publications:
Selected websites
The Max Planck Institute
The Max Planck Institute (MPI) for Psycholinguistics is an institute of the German Max Planck Society. Its mission is to undertake basic research into the psychological, social and biological foundations of language. The Language and Genetics department was newly established at the MPI in 2010, with the goal of using genes as windows into the biological basis of speech and language. The MPI is situated on the campus of the Radboud University in Nijmegen. It participates in the Donders Institute for Brain, Cognition and Behaviour, as well as the Centre for Language Studies, hosting a joint graduate school, the International Max Planck Research School (IMPRS) in Language Sciences.
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


