Coelacanth genome surfaces
An international team of researchers has decoded the genome of a creature whose evolutionary history is both enigmatic and illuminating: the African coelacanth. A sea-cave dwelling, five-foot-long fish with limb-like fins, the coelacanth was once thought to be extinct.
The team’s study of the coelacanth genome confirms what many scientists had long suspected: genes in coelacanths are evolving more slowly than in other organisms. They also find intriguing suggestions of regions of the genome that helped in the move from sea to land.
A living coelacanth was discovered off the African coast in 1938 and, since then, the evolutionary position of these ancient-looking fish – popularly known as ‘living fossils’ – has been an enigma. Coelacanths today closely resemble the fossilised skeletons of their more than 300-million-year-old ancestors.
“We found that the genes overall are evolving significantly more slowly in the coelacanth than in every other fish and land vertebrate that we looked at. This is the first time that we’ve had a big enough gene set to really see that.”
Jessica Alföldi A research scientist at the Broad Institute and co-first author of the paper on the coelacanth genome, which appears in Nature
Researchers hypothesize that this slow rate of change may be because coelacanths simply have not needed to change: they live primarily off of the Eastern African coast (a second coelacanth species lives off the coast of Indonesia), at ocean depths where relatively little has changed over the millennia.
“We often talk about how species have changed over time. But there are still a few places on Earth where organisms don’t have to change, and this is one of them. Coelacanths are likely very specialized to such a specific, non-changing, extreme environment – it is ideally suited to the deep sea just the way it is.”
Kerstin Lindblad-Toh Scientific director of the Broad Institute’s vertebrate genome biology group and senior author
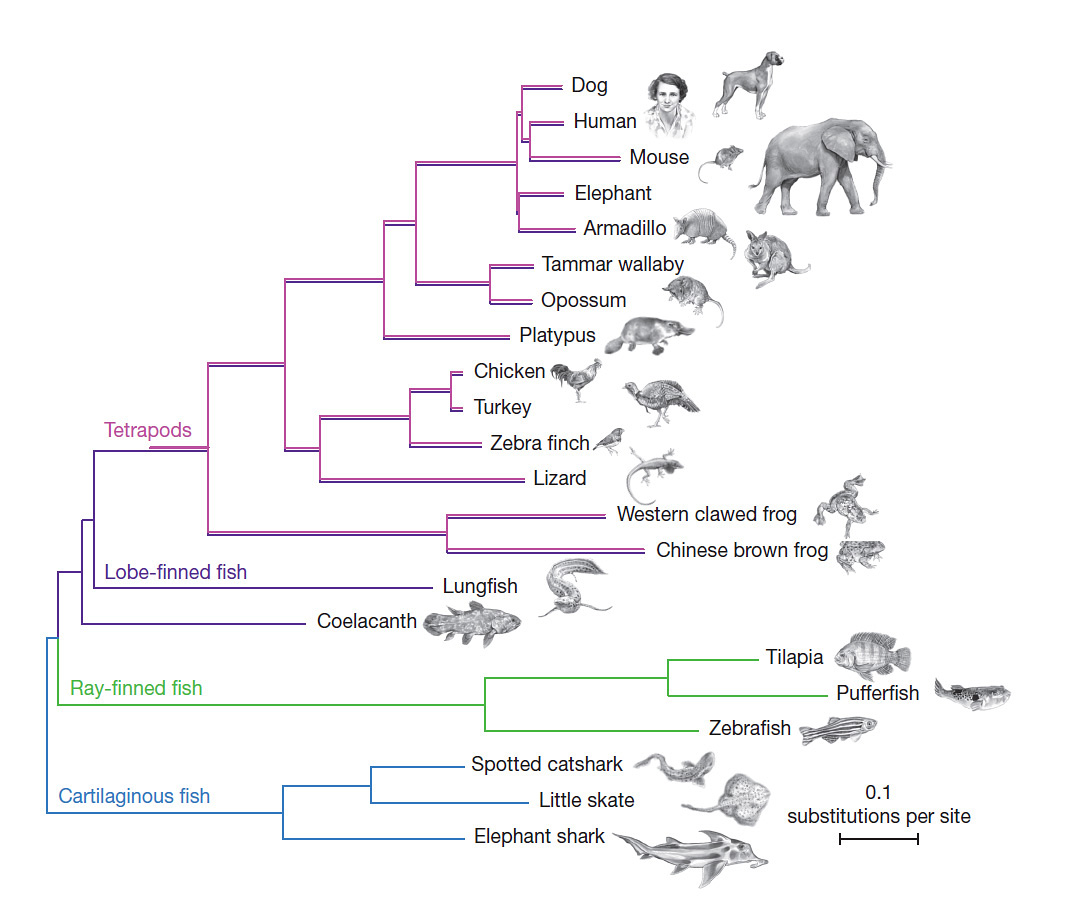
The coelacanth genome has also allowed scientists to test other long-debated questions. For example, coelacanths possess some features that look oddly similar to those seen only in animals that dwell on land, including ‘lobed’ fins, which resemble the limbs of four-legged land animals (known as tetrapods). Another odd-looking group of fish known as lungfish possesses lobed fins too. The genomes of these species suggest that the lungfish is more closely related to the tetrapod – and ultimately the human – ancestor.
A genome is not a dead and dry set of instructions, but a lens into the past and into the future. We can glimpse the past and the distance travelled to today only because of our careful gene descriptions, our annotation. The gene structures we describe are the genomic skeleton for this ‘living-fossil’ fish, bearing marks of past history and presaging new developments in evolution.
“Through these gene descriptions, we see hints of the march from sea to land that occurred perhaps 400 million years ago.”
Dr Steve Searle from the Wellcome Trust Sanger Institute
Because of their resemblance to fossils dating back millions of years Coelacanths today are often referred to as ‘living fossils’ – a term coined by Charles Darwin. But the coelacanth is not a relic of the past brought back to life: it is a species that has survived, reproduced, but changed very little in appearance for millions of years.
However, the coelacanth is still a critical organism to study in order to understand what is often called the water-to-land transition. Lungfish may be more closely related to land animals, but its genome remains inscrutable: at 100 billion genetic letters in length, the lungfish genome is simply too unwieldy for scientists to sequence, assemble, and analyse. The coelacanth’s more modest-sized genome (comparable in length to our own) is yielding valuable clues about the genetic changes that may have allowed tetrapods to flourish on land.
The researchers made several unusual discoveries by looking at which genes were lost when vertebrates came on land as well as changes to regulatory elements that govern where, when, and to what extent genes are active. For example, they found changes to regulatory regions controlling genes involved in sense of smell, the recruitment of several regulatory regions that were used by tetrapods to drive limb development and changes to a gene that specifies a key protein involved in the urea cycle – part of the excretion process that land animals use, whereas fish do not.
“This is just the beginning of many analyses on what the coelacanth can teach us about the emergence of land vertebrates, including humans, and, combined with modern empirical approaches, can lend insights into the mechanisms that have contributed to major evolutionary innovations.”
Chris Amemiya, a member of the Benaroya Research Institute, professor at the University of Washington and co-first author of the paper
Coelacanths are an endangered species, meaning that each sample obtained was precious. But the difficulties in obtaining a sample and the challenges of sequencing it also knitted the community together.
“The international nature of the work, its evolutionary value and history, and the fact that it was a technically challenging project really brought people together. We had representatives from every populated continent on earth working on this project.”
Kerstin Lindblad-Toh The Broad Institute
Although its genome offers some tantalising answers, the research team anticipates that further study of the fish’s immunity, respiration, physiology, and more will lead to deep insights into how some vertebrates adapted to life on land, while others remained creatures of the sea.
More information
Funding
This work was supported by the African Coelacanth Ecosystem Programme of the South African National Department of Science and Technology, the National Human Genome Research Institute (NHGRI), the European Science Foundation. Additional contributions were made by the Genomics Sequencing Platform of the Broad Institute, the Association pour le Protection de Gombesa and other agencies.
A full list of funding can be found in the paper on the Nature website.
Participating Centres
A full list of participating centres can be found in the paper on the Nature website.
Publications:
Selected websites
Broad Institute of Harvard and MIT
The Eli and Edythe L. Broad Institute of Harvard and MIT was launched in 2004 to empower this generation of creative scientists to transform medicine. The Broad Institute seeks to describe all the molecular components of life and their connections; discover the molecular basis of major human diseases; develop effective new approaches to diagnostics and therapeutics; and disseminate discoveries, tools, methods and data openly to the entire scientific community.
Founded by MIT, Harvard and its affiliated hospitals, and the visionary Los Angeles philanthropists Eli and Edythe L. Broad, the Broad Institute includes faculty, professional staff and students from throughout the MIT and Harvard biomedical research communities and beyond, with collaborations spanning over a hundred private and public institutions in more than 40 countries worldwide.
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


