Genetics of flu susceptibility
A genetic finding could help explain why influenza becomes a life-threatening disease to some people while it has only mild effects in others. New research led by the Wellcome Trust Sanger Institute has identified for the first time a human gene that influences how we respond to influenza infection.
People who carry a particular variant of a gene called IFITM3 are significantly more likely to be hospitalised when they fall ill with influenza than those who carry other variants, the team found. This gene plays a critical role in protecting the body against infection with influenza and a rare version of it appears to make people more susceptible to severe forms of the disease. The results are published in the journal Nature.
A central question about viruses is why some people suffer badly from an infection and others do not. IFITM3 is an important protein that protects cells against virus infection and is thought to play a critical role in the immune system’s response against such viruses as H1N1 pandemic influenza, commonly known as ‘swine flu’. When the protein is present in large quantities, the spread of the virus in lungs is hindered, but if the protein is defective or absent, the virus can spread more easily, causing severe disease.
“Although this protein is extremely important in limiting the spread of viruses in cells, little is known about how it works in lungs,” explains Aaron Everitt, first author from the Wellcome Trust Sanger Institute. “Our research plays a fundamental part in explaining how both the gene and protein are linked to viral susceptibility.”
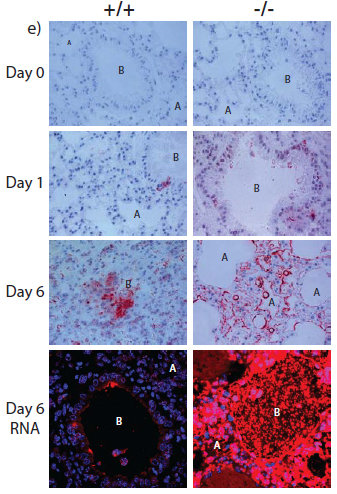
The antiviral role of IFITM3 in humans was first suggested by studies using a genetic screen, which showed that the protein blocked the growth of influenza virus and dengue virus in cells. This led the team to ask whether IFITM3 protected mice from viral infections. They removed the IFITM3 gene in mice and found that once they contracted influenza, the symptoms became much more severe compared to mice with IFITM3. In effect, they found the loss of this single gene in mice can turn a mild case of influenza into a fatal infection.
The researchers then sequenced the IFITM3 genes of 53 patients hospitalised with influenza and found that some have a genetic mutant form of IFITM3, which is rare in normal people. This variant alters the IFITM3 gene and makes cells more susceptible to viral infection.
“Since IFITM3 appears to be a first line defender against infection, our efforts suggest that individuals and populations with less IFITM3 activity may be at increased risk during a pandemic and that IFITM3 could be vital for defending human populations against other viruses such as avian influenza virus and dengue virus.”
Dr. Abraham Brass Co-senior author and Assistant Professor at the Ragon Institute of MGH, MIT and Harvard and the Gastrointestinal Unit of Massachusetts General Hospital
This research was a collaboration between institutes in the United States and the United Kingdom. The samples for this study were obtained from the MOSAIC consortium in England and Scotland, co-ordinated from the Centre for Respiratory Infection (CRI) at Imperial College London, and the GenISIS consortium in Scotland at the Roslin Institute of University of Edinburgh. These were pivotal for the human genetics component of the work.
“Collectively, these data reveal that the action of a single antiviral protein, IFITM3, can profoundly alter the course of the flu and potentially other viruses in both human and mouse. To fully understand how both the protein and gene control our susceptibility to viral infections, we need to study the mechanisms of the gene variant more closely.
“Our research is important for people who have this variant as we predict their immune defences could be weakened to some virus infections. Ultimately as we learn more about the genetics of susceptibility to viruses, then people can take informed precautions, such as vaccination to prevent infection.”
Professor Paul Kellam Co-senior author from the Wellcome Trust Sanger Institute
“During the recent swine flu pandemic, many people found it remarkable that the same virus could provoke only mild symptoms in most people, while, more rarely, threatening the lives of others. This discovery points to a piece of the explanation: genetic variations affect the way in which different people respond to infection.
“This important research adds to a growing scientific understanding that genetic factors affect the course of disease in more than one way. Genetic variations in a virus can increase its virulence, but genetic variations in that virus’s host – us – matter greatly as well.”
Sir Mark Walport Director of the Wellcome Trust
More information
Additional quotes
“This new discovery is the first clue from our detailed study of the devastating effects of flu in hospitalised patients. It vindicates our conviction that there is something unusual about these patients, and that ground-breaking clinical studies can be performed in the UK.”
Professor Peter Openshaw Director of the Centre for Respiratory Infection (CRI) at Imperial College London
“This work contributes greatly to our understanding of the reasons why susceptibility to serious infectious disease is so highly heritable. It is a study that couldn’t have happened without the contribution of the patients of the GenISIS consortium in Scotland and the MOSAIC consortium in England and Scotland. We are extremely grateful for their support.”
Professor David Hume Director of The Roslin Institute and author on the study
Funding
This work was supported by the Wellcome Trust. The MOSAIC work was supported by Imperial’s National Institute for Health Research Comprehensive Biomedical Research Centre (cBRC) , the Wellcome Trust and Medical Research Council UK. The GenISIS work was supported by the Chief Scientist Office (Scotland) and the Roslin Institute of the University of Edinburgh. A.L.B. is the recipient of a Charles H. Hood Foundation Child Health Research Award, and is supported by grants from the Phillip T. and Susan M. Ragon Institute Foundation, the Bill and Melinda Gates Foundation’s Global Health Program and the National Institute of Allergy and Infectious Diseases. J.K.B. is supported by a Wellcome Trust Clinical Lectureship through the Edinburgh Clinical Academic Track (ECAT).
Participating Centres
A full list of participating centres can be found at the Nature website.
Publications:
Selected websites
Imperial College London
Imperial College London consistently rated amongst the world’s best universities, Imperial College London is a science-based institution with a reputation for excellence in teaching and research that attracts 14,000 students and 6,000 staff of the highest international quality. Innovative research at the College explores the interface between science, medicine, engineering and business, delivering practical solutions that improve quality of life and the environment – underpinned by a dynamic enterprise culture.
Since its foundation in 1907, Imperial’s contributions to society have included the discovery of penicillin, the development of holography and the foundations of fibre optics. This commitment to the application of research for the benefit of all continues today, with current focuses including interdisciplinary collaborations to improve global health, tackle climate change, develop sustainable sources of energy and address security challenges.
In 2007, Imperial College London and Imperial College Healthcare NHS Trust formed the UK’s first Academic Health Science Centre. This unique partnership aims to improve the quality of life of patients and populations by taking new discoveries and translating them into new therapies as quickly as possible.
The Roslin Institute
The Roslin Institute at the University of Edinburgh received a total of £8.5M investment from the Biotechnology and Biological Sciences Research Council in 2010-11. The Institute undertakes research within the framework of BBSRC Institute Strategic Programmes focussed on the health and welfare of animals, and applications of basic animal sciences in human and veterinary medicine, the livestock industry and food security.
The Ragon Institute of MGH, MIT and Harvard
The Ragon Institute of MGH, MIT and Harvard was established in 2009 with a gift from the Phillip T. and Susan M. Ragon Foundation, creating a collaborative scientific mission among these institutions to harness the immune system to combat and cure human diseases. The primary initial focus of the institute is to contribute to the development of an effective AIDS vaccine. Administratively based at Massachusetts General Hospital, the Ragon Institute draws scientists and engineers from diverse backgrounds and areas of expertise across the Harvard and MIT communities and throughout the world, in order to apply the full arsenal of scientific knowledge to understanding mechanisms of immune control and immune failure and to apply these advances to directly benefit patients.
Massachusetts General Hospital
Massachusetts General Hospital, founded in 1811, is the original and largest teaching hospital of Harvard Medical School. The MGH conducts the largest hospital-based research program in the United States, with an annual research budget of more than $750 million and major research centers in AIDS, cardiovascular research, cancer, computational and integrative biology, cutaneous biology, human genetics, medical imaging, neurodegenerative disorders, regenerative medicine, reproductive biology, systems biology, transplantation biology and photomedicine.
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


