Tracing the UK's No1 sexually transmitted infection
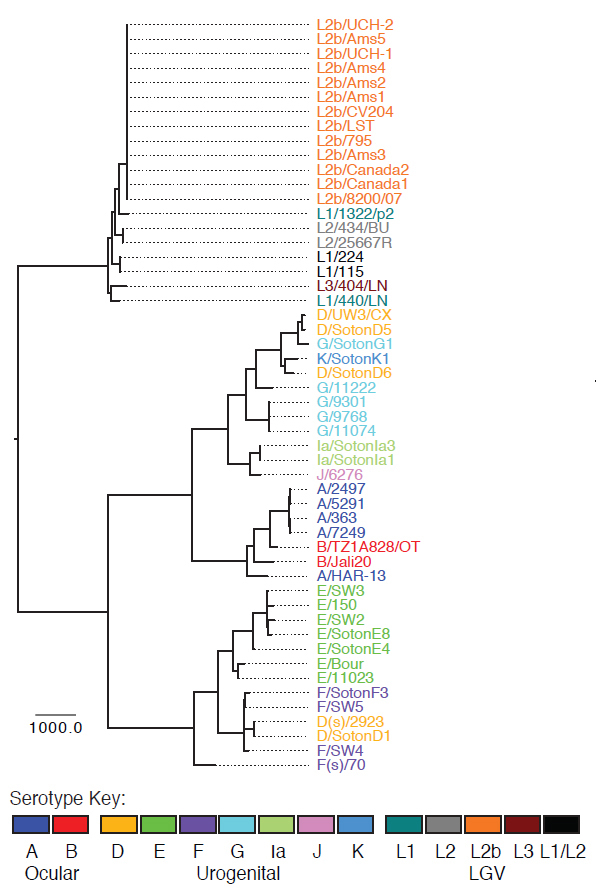
In a study released today in Nature Genetics, researchers have found that Chlamydia has evolved more actively than was previously thought. Using whole-genome sequencing the researchers show that the exchange of DNA between different strains of Chlamydia to form new strains is much more common than expected.
The team highlights that current clinical testing methods cannot capture the variation between Chlamydia strains. Changes to the genome structure are missed by current tests for Chlamydia, giving us the false impression that Chlamydia hardly changes at all. The researchers are working with hospitals to use their results to improve Chlamydia testing.
Chlamydia trachomatis is the most common sexually transmitted infection (STI) both in the UK and globally, with approximately 100 million new cases worldwide each year. It is also the most common cause of infectious blindness, or trachoma, in the developing world. Relatively little is known about the evolution of the different strains of Chlamydia that are causing infection.
“Scientists recently discovered that if two Chlamydia strains co-infect the same person at the same time, they can swap DNA by a process called recombination. This was originally thought only to affect a few ‘hotspots’ within the genome. We were very surprised to find recombination is far more widespread than previously thought.”
Dr Simon Harris Lead author from the Wellcome Trust Sanger Institute
The team found there appeared to be no barriers to the swapping of DNA when circumstances allow, even between bacterial strains associated with infecting different parts of the body.
“Despite this being the most important sexually transmitted infection in the world, until now it’s clear that there are major gaps in our knowledge of the strains that are currently circulating, their evolution and natural history.”
Professor Ian Clarke Senior author from the University of Southampton, Faculty of Medicine
Clinical diagnosis of Chlamydia infections simply returns a positive or negative result, providing no information about the nature of the infecting strain. This makes it impossible to determine whether a person who tests positive again after antibiotic treatment has picked up a second infection or if their treatment has failed. The significance of this is that although antibiotic-resistant Chlamydia has never been seen in patients, it can occur in the laboratory. If resistance did occur in the general population, it would not be detected by current diagnostic procedures.
“Until now a person treated with antibiotics with a reoccurring infection of C. trachomatis was assumed to have been re-infected. The current gaps in our understanding of the population makeup of Chlamydia limit our ability to implement health policies, because we do not fully understand how Chlamydia spreads within our population.”
Dr Nicholas Thomson Senior author from the Wellcome Trust Sanger Institute
This study is not just important for the treatment of the sexually transmitted strains of Chlamydia but also for the treatment of African Chlamydia strains that can cause infectious blindness, or trachoma. The team sequenced strains of African Chlamydia and found that these strains are also using recombination to fool the human immune system.
“For many years various groups have observed co-circulating strains of Chlamydia causing trachoma. In our study we have shown that some strains appear to have swapped only their surface coat. This provides real clues as to how this bacterium is able to avoid the human immune system and cause disease.”
Dr Martin Holland from the London School of Hygiene & Tropical Medicine
This study shows that whole genome sequencing is the resolution required to understand in fine detail the spread of Chlamydia through a population. Scientists at the Sanger Institute are currently working with hospitals to bring this technology into the clinical setting. The challenge that researchers face is to provide enough DNA from routine samples to generate a whole genome sequence.
More information
Funding
This work was funded by the Wellcome Trust.
Participating Centres
A full list of participating centres can be found at the Nature Genetics website.
Publications:
Selected websites
The University of Southampton
The University of Southampton is a leading UK teaching and research institution with a global reputation for leading-edge research and scholarship across a wide range of subjects in engineering, science, social sciences, health and humanities. With over 23,000 students, around 5000 staff, and an annual turnover well in excess of £435 million, the University of Southampton is acknowledged as one of the country’s top institutions for engineering, computer science and medicine. We combine academic excellence with an innovative and entrepreneurial approach to research, supporting a culture that engages and challenges students and staff in their pursuit of learning. The University is also home to a number of world-leading research centers including the Institute of Sound and Vibration Research, the Optoelectronics Research Centre, the Web Science Trust and Doctoral training Centre, the Centre for the Developmental Origins of Health and Disease, the Southampton Statistical Sciences Research Institute and is a partner of the National Oceanography Centre at the Southampton waterfront campus.
The London School of Hygiene & Tropical Medicine
The London School of Hygiene & Tropical Medicine is a world-leading centre for research and postgraduate education in public and global health, with 4000 students and more than 1300 staff working in over 100 countries. The School is one of the highest-rated research institutions in the UK, and was recently cited as one of the world’s top universities for collaborative research. The School’s mission is to improve health and health equity in the UK and worldwide; working in partnership to achieve excellence in public and global health research, education and translation of knowledge into policy and practice.
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


