Genetic cause for hearing impairment discovered
Researchers have used the power of next-generation sequencing to identify a gene involved in hearing impairment. These findings reveal a new and unexpected function for a previously well-studied gene.
The study recently released, found that this mutation is involved in otitis media, an inflammation of the middle ear, which is a prominent cause of hearing impairment especially in children. This research highlights the power of genome sequencing to identify causative mutations.
“Otitis media is the leading cause of surgery in children in the developed world. It affects 35 per cent of children under the age of 2 in Europe. With these expanding developments in the genetics and the understanding of otitis media, there is a possibility of reducing the need for invasive surgery in young children in the future.”
Jennifer Hilton from the Wellcome Trust Sanger Institute and first author on the paper
This research emphasizes two important concepts. Firstly, exome sequencing can detect mutations that increase the chances of developing a disease but don’t necessarily cause the problem in every person carrying the mutation (known as reduced penetrance). Secondly, the middle ear disease was detected only because the researchers looked for it, and could easily be missed in a mouse, suggesting that we only find what we look for in mouse models of disease.
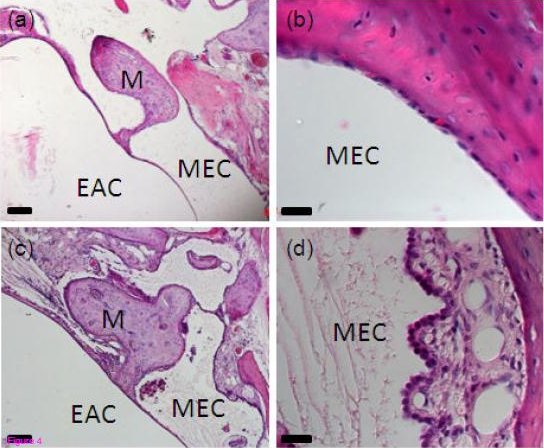
The team studied the exome sequence of one affected mouse and found a mutation in the Islet1 gene. They then looked for this mutation in a collection of affected mice and found that all contained the mutation in the Islet1 gene. The mutants show a thickened and inflamed lining of the middle ear and excessive fluid and cell debris in what should be an air-filled middle ear cavity, which is a clear indication of chronic otitis media.
Although the Islet1 gene has been extensively studied, its role in hearing was previously unrecognised. The team used exome sequencing to selectively decode the gene rich regions of the genome. Because it sequences all genes in a genome, exome sequencing gives an unbiased view of genes associated with rare or common disorders.
“Our work reflects the power of exome sequencing to identify mutations that have less obvious traits. This is the first time that exome sequencing has revealed a mutation associated with deafness in a case where the low penetrance of the trait meant that we could not map the mutation to a specific chromosome but had to look at the whole genome.”
Karen P Steel senior author from the Wellcome Trust Sanger Institute
“Previous heritability studies suggested that otitis media has a significant genetic component. Mouse mutants have played a key role in identifying genes predisposing to otitis media. Our research team has shown that a new genetic factor is involved With the discovery of the mutation in the Islet1 gene.”
Morag Lewis, from the Wellcome Trust Sanger Institute and second author on the paper
This mutation remained elusive for many years, as it does not have any immediately apparent physical traits, and in this way resembles many variants in the human genome. The type of sequencing used by the team to detect the mutation that predisposes the carriers to otitis media highlights the opportunities for use of this technology in the future.
More information
Funding
This work was supported by the Wellcome Trust (grant 077189), the EC (CT97-2715), and the MRC.
Participating Centres
- Wellcome Trust Sanger Institute, Hinxton, Cambridge CB10 1SA, UK
- Current address: NIH, NIDCD, Bethesda, MD 20892, USA
- EMBL European Bioinformatics Institute, Hinxton, Cambridge CB10 1SD, UK
Publications:
Selected websites
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


