Real-time genomics applied to fatal outbreak
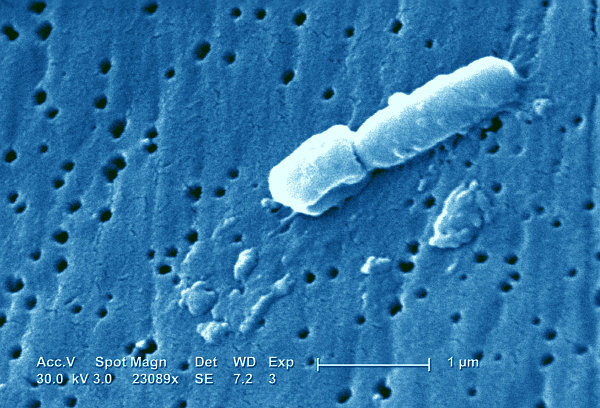
Two scientists from the Wellcome Trust Sanger Institute have been helping to identify the strain of a highly antibiotic-resistant bacterium at a Dutch hospital which infected more than 80 patients, of which 27 have died. It is the first time that real-time genomics has been applied for diagnostics during an outbreak.
Drs Nick Thomson and Tom Connor were part of an international collaboration asked to contain the outbreak of Klebsiella pneumoniae Oxa-48 by the Dutch National Institute for Public Health and the Environment (RIVM). More than 2,000 people at Maasstad Hospital, Rotterdam were also at risk of infection.
Experts at RIVM teamed up with scientists of the Medical Faculty of the University Münster, Germany, bioinformaticians from Life Technologies Corporation in America, and scientists from the Sanger Institute in Cambridgeshire, to develop a highly specific and rapid molecular screening test to detect and prevent further spread of the multiresistant organism.
The Sanger Institute researchers were asked to collaborate because of their unique global database of Klebsiella pneumonia.
“Tom and I have worked on many infections before and we have a unique global database of Klebsiella pneumoniae which helped us identify the specific strain which was causing the problems in the hospital,. The real significance of what Tom and the team have managed to do was to identify the distinguishing features of the outbreak strain that could be used to develop a rapid test. What they also found out was that this wasn’t a novel strain, as a similar strain had also been identified in Vietnam.”
Dr Nick Thomson from the Pathogen Genomics team at the Wellcome Trust Sanger Institute
Dr Connor put his other projects on hold to help the Dutch colleagues as a matter of urgency.
“It is the first time anything like this has happened to us. At the moment we are only just getting the resources together to work collaboratively like this, and that’s very exciting. In a few years, most of the bacterial pathogens we work on will have these global databases of strains which will enable scientists around the world to replicate this work in other outbreak situations.”
Dr Tom Connor Wellcome Trust Sanger Institute
“It was essential to bring together quickly the right people and resources so that we were able to respond to the potential spread of this multi-resistant bacterium among patients in Dutch hospitals. We are especially pleased about the role of rapid whole genome sequencing of the outbreak strain.”
Professor Dr Hajo Grundmann Epidemiologist working at the RIVM and the University Medical Centre Groningen, who coordinated the joint task force
“When we first used the PGM during the German EHEC 2011 outbreak, this was just a proof of principle. However, this time the rapid bacterial whole genome sequencing will make a difference for the protection of patients’ health. The sequence data laid the ground for the development of a specific molecular test. That is a true first real-time ‘genomics for diagnostics’ application.”
Microbiologist Professor Dr Dag Harmsen from the Department of Periodontology at the University Münster, who led the team responsible for sequencing on an Ion TorrentPGM
Bioinformaticans from Life Technologies used the data from a single Ion 316 chip to assemble a draft genome of the K. pneumoniae Oxa-48 outbreak strain (isolate 1191100241), which is available to the public through the National Center for Biotechnology Information.
“Using our TaqMan® assay development pipeline, we were able to compare this genome rapidly to other publicly available Klebsiella genomes and identify 36 candidate signature sequences that could be used to develop an outbreak strain-specific multiplex PCR test.”
Craig Cummings from Life Technologies
By comparing these candidate signature sequences against the publically available data for more than 200 additional Klebsiella genomes from an ongoing worldwide population study Tom and Nick identified two regions that remain entirely specific for the Dutch hospital outbreak strain. PCR assays targeting these two unique sequences together with the antibiotic resistance genes were then evaluated and field-tested by Grundmann’s team at the RIVM to develop a multiplex molecular screening test.
“Overall, this is a good example of how rapid next generation sequencing can help controlling outbreaks and how joint forces from different countries and continents can achieve quick results to prevent further infections for the sake of the Dutch hospital patients.”
Professor Dr Hajo Grundmann, RIVM and the University Medical Centre Groningen
More information
Resources available
NCBI/SRA accession number: SRA043951
Selected websites
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute, which receives the majority of its funding from the Wellcome Trust, was founded in 1992. The Institute is responsible for the completion of the sequence of approximately one-third of the human genome as well as genomes of model organisms and more than 90 pathogen genomes. In October 2006, new funding was awarded by the Wellcome Trust to exploit the wealth of genome data now available to answer important questions about health and disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


