New elegant technique used for genomic archaeology
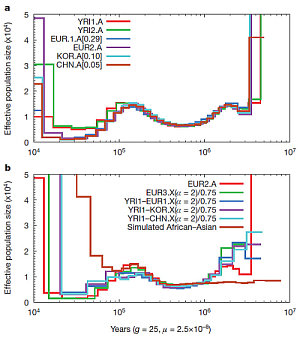
Researchers have probed deeper into human evolution by developing an elegant new technique to analyse whole genomes from different populations. One key finding from the Wellcome Trust Sanger Institute’s study is that African and non-African populations continued to exchange genetic material well after migration out of Africa 60,000 years ago. This shows that interbreeding between these groups continued long after the original exodus.
For the first time genomic archaeologists are able to infer population size and history using single genomes, a technique that makes fewer assumptions than existing methods, allowing for more detailed insights. It provides a fresh view of the history of mankind from 10,000 to one million years ago.
“Using this algorithm, we were able to provide new insights into our human history. First, we see an apparent increase in effective human population numbers around the time that modern humans arose in Africa over 100,000 years ago.
“Second, when we look at non-African individuals from Europe and East Asia, we see a shared history of a dramatic reduction in population, or bottleneck, starting about 60,000 years ago, as others have also observed. But, unlike previous studies, we also see evidence for continuing genetic exchange with African populations for tens of thousands of years after the initial out-of-Africa bottleneck until 20,000 to 40,000 years ago.
“Previous methods to explore these questions using genetic data have looked at a subset of the human genome. Our new approach uses the whole sequence of single individuals, and relies on fewer assumptions. Using such techniques we will be able to capitalize on the revolution in genome sequencing and analysis from projects such as The 1000 Genomes Project and, as more people are sequenced, build a progressively finer detail picture of human genetic history.”
Dr Richard Durbin Joint head of Human Genetics and leader of the Genome Informatics Group at the Sanger Institute
The team sequenced and compared four male genomes: one each from China, Europe, Korea and West Africa respectively. The researchers found that, although the African and non-African populations might have started to differentiate as early as 100,000 to 120,000 years ago, they largely remained as one population until approximately 60,000 to 80,000 years ago.
Following this the European and East Asian ancestors went through a period where their effective population size crashed to approximately one-tenth of its earlier size, overlapping the period when modern human fossils and artefacts start to appear across Europe and Asia. But, for at least the first 20,000 years of this period, it appears that the out-of-Africa and African populations were not genetically separated. A possible explanation could be that new emigrants from Africa continued to join the out-of-Africa populations long after the original exodus.
“This elegant tool provides opportunities for further research to enable us to learn more about population history. Each human genome contains information from the mother and the father, and the differences between these at any place in the genome carry information about its history. Since the genome sequence is so large, we can combine the information from tens of thousands of different places in the genome to build up a composite history of the ancestral contributions to the particular individual who was sequenced.
“We can also get at the historical relationship between two different ancestral populations by comparing the X chromosomes from two males. This works because men only have one copy of the X chromosome each, so we can combine the X chromosomes of two men and treat them in the same way as the rest of the genome for one person, with the results telling us about the way in which the ancestral populations of the two men separated.
“The novel statistical method we developed is computationally efficient and doesn’t make restrictive assumptions about the way that population size changed. Although not inconsistent with previous results, these findings allow new types of historical events to be explored, leading to new observations about the history of mankind.”
Heng Li Co-author of the study from the Sanger Institute
The researchers believe that this technique can be developed further to enable even more fine-grained discoveries by sequencing multiple genomes from different populations. In addition, beyond human history, there is also the potential to investigate the population size history of other species for which a single genome sequence has been obtained.
More information
Funding
This work was supported by the Wellcome Trust.
Publications:
Selected websites
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute, which receives the majority of its funding from the Wellcome Trust, was founded in 1992. The Institute is responsible for the completion of the sequence of approximately one-third of the human genome as well as genomes of model organisms and more than 90 pathogen genomes. In October 2006, new funding was awarded by the Wellcome Trust to exploit the wealth of genome data now available to answer important questions about health and disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


