Movies for the human genome
The data have been developed using the freely available human genome sequence: as with the public release of that sequence, the consortium that produced the current results has made them freely available to the research community.
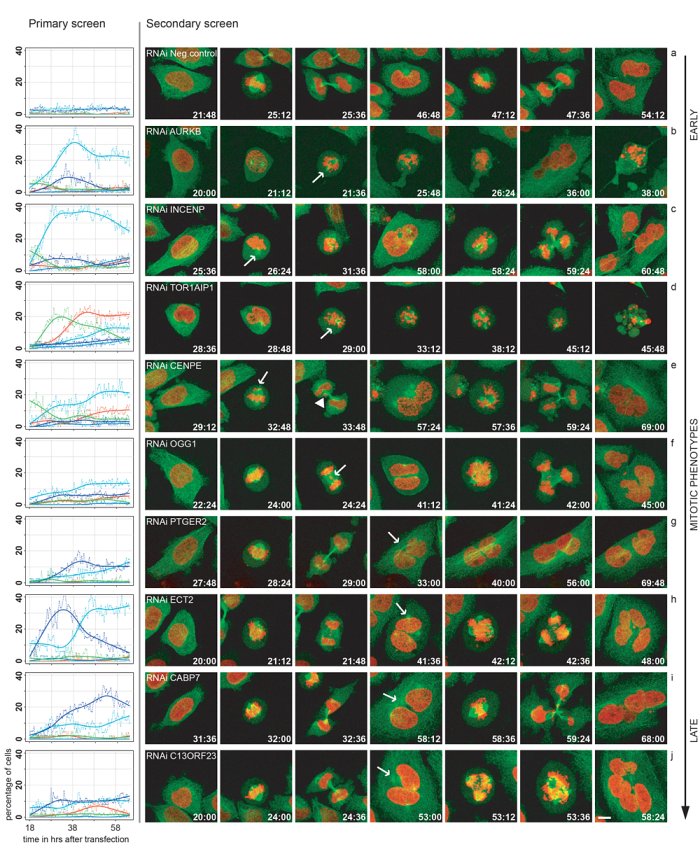
The Mitocheck Consortium developed high-throughput systems to disrupt the working of each gene and to study the consequences for cell behaviour using time-lapse microscopy: this ‘Candid Camera’ of the cell caught how cells respond to gene disruption.
The team produced about 190,000 movies.
“This work is the fruit of an innovative European collaboration in functional genomics, bringing multiple large-scale experimental strategies to bear on a key component of basic biology: how cells manage the doubling of their DNA when they divide. Because of its scale it would not have been possible without new software and analysis tools.”
Dr Richard Durbin A researcher at the Wellcome Trust Sanger Institute
To carry out biology on this scale, the team developed automated systems to inhibit gene function and to study the effects on cell behaviour such as migration, division. The observations were supported by newly developed computer programmes to report the cell observations.
The team studied almost two billion cell nuclei.
Cell division is often poorly captured in studies of mutation, in part simply because it is such a short part of a cell’s life. However this vital part of a cell’s – and our – life is ideally captured by time-lapse microscopy. The study found almost 600 genes that could be involved in these basic functions.
The system uses reduction in gene activity caused by siRNAs – small, interfering RNAs designed to match and thus interfere with the sequence of one specific gene. The elegant high throughput platform that EMBL scientists developed to silence all of an organism’s genes in a fast and systematic manner is itself proving a boon to the scientific community.
“A year after we developed these new siRNA microarrays they’re already in use by over 10 research groups from across Europe.”
Dr Rainer Pepperkok who led the method’s development at EMBL
The team have yet to uncover exactly how these genes act at the molecular level – a task which will be tackled by a follow-up project called MitoSys. All data from this follow-up work will also be made freely available online, creating what Professor Ellenberg describes as a ‘one-stop-shop’ for mitosis research.
“Without mitosis, nothing happens in life, really and when mitosis goes wrong, you get defects like cancer.”
Jan Ellenberg who led the study at EMBL
More information
Data availability
Data are available from the Mitocheck consortium as a resource to the community at http://www.mitocheck.org.
Funding
This project was funded by grants by the European Commission, by the German Federal Ministry of Education and Research (BMBF) in the framework of the National Genome Research Network, by the Landesstiftung Baden Wuerttemberg and by the Wellcome Trust.
Participating Centres
- MitoCheck Project Group, Gene Expression and Cell Biology/Biophysics Units, Structural and Computational Biology Unit, European Molecular Biology Laboratory (EMBL), Heidelberg, Germany
- Max Planck Institute for Molecular Cell Biology and Genetics, Dresden, Germany
- ETH Zurich, Institute of Biochemistry, ETH-Hoenggerberg, Zurich, Switzerland
- Leica Microsystems CMS GmbH, Mannheim, Germany
- European Bioinformatics Institute, European Molecular Biology Laboratory, Cambridge, UK
- Division of Theoretical Bioinformatics, German Cancer Research Center, Heidelberg, Germany
- Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK
- Institute for Molecular Pathology, Vienna, Austria.
Publications:
Selected websites
The European Molecular Biology Laboratory
The European Molecular Biology Laboratory is a basic research institute funded by public research monies from 20 (Austria, Belgium, Croatia, Denmark, Finland, France, Germany, Greece, Iceland, Ireland, Israel, Italy, Luxembourg, Norway, Portugal, Spain, Sweden, Switzerland and the United Kingdom) and associate member state Australia. Research conducted by approximately 85 independent groups covering the spectrum of molecular biology. The Laboratory has five Laboratory in Heidelberg, and Outstations in Hinxton (the European Bioinformatics Institute), Grenoble, Hamburg, and near Rome. The cornerstones of EMBL’s mission are: to perform basic research in molecular biology; to train scientists, visitors at all levels; to offer vital services to scientists in the member states; to develop new instruments and methods in and to actively engage in technology transfer activities. EMBL’s International PhD Programme has a student body of Laboratory also sponsors an active Science and Society programme. Visitors from the press and public are welcome.
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute, which receives the majority of its funding from the Wellcome Trust, was founded in 1992. The Institute is responsible for the completion of the sequence of approximately one-third of the human genome as well as genomes of model organisms and more than 90 pathogen genomes. In October 2006, new funding was awarded by the Wellcome Trust to exploit the wealth of genome data now available to answer important questions about health and disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


