Exploring genetic effects in cells
Researchers report the most extensive and comprehensive study of the impact of genetic variations on gene activity. The study uses RNA sequencing at an unprecedented resolution to reveal genetic effects in cellular processes.
The team used novel technologies to obtain a detailed picture of how gene activity in blood cells differs among people and to describe how variations in DNA are responsible for these differences. The results have important implications for understanding genetic susceptibility to common and rare diseases as well as the basis of natural variation between people.
“For the first time we are able to ‘read’ the sequence of almost all the RNA molecules in the cell and compare them among individuals. As a computational biologist and geneticist, this is a dream come true.”
Dr Stephen Montgomery Lead author of the study, formerly of the Wellcome Trust Sanger Institute and now at the University of Geneva
Genetic activity is based largely on the number of ‘messenger’ RNA copies of a gene that a cell produces: the messengers contain instructions to build proteins. More messengers, more protein.
Most often, levels of RNA messengers were estimated from DNA chips called arrays. The team show the new method is as good as or better than array methods. Most important, the new method can distinguish slightly different versions of messengers invisible to the array methods. Different messengers might produce different proteins.
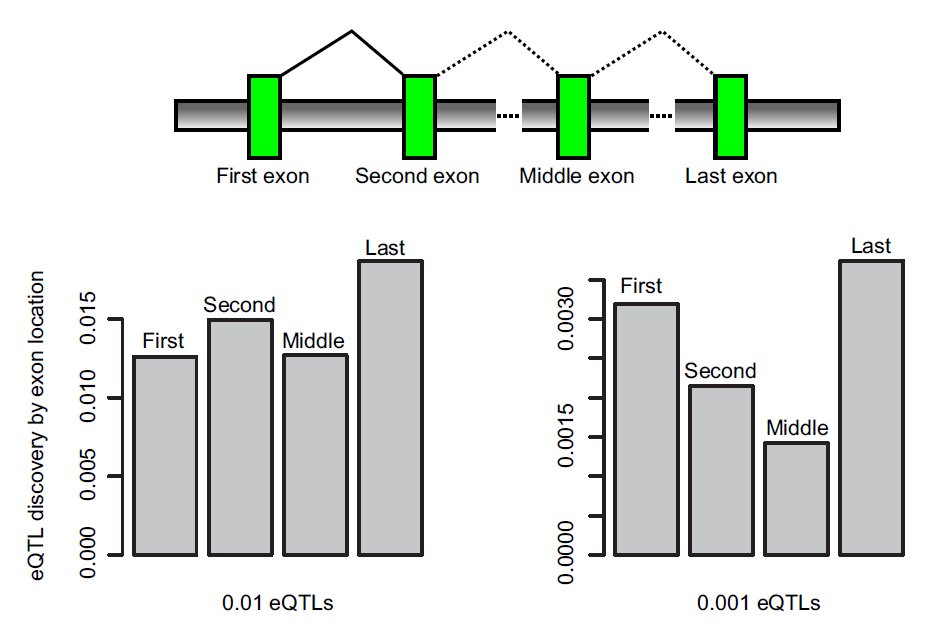
The new method produces massive amounts of data and the team had to develop new software called FluxCapacitor to tease apart the data sets and to identify alternative messengers.
Previous studies have told us about rough individual differences in the quantity of RNA from each gene in the cell. But this is only the tip of the iceberg in terms of defining the molecular consequences. In this study, conducted using blood cells of 60 individuals of European descent, the investigators have been able to describe in detail the molecular differences in RNA among individuals.
The ability to read the RNA sequence in so many individuals is of unprecedented scale and brings the understanding of genetic variation to a new level.
“This is a remarkable development that solves four major challenges in linking genetic changes to messenger levels – the heart of gene activity. We show that our method can find more genetic variants than arrays, we show that we can work with relatively small sample numbers and still get consistent results, we show that we can find rare variants that affect gene regulation and, for the first time, we show that we can discover genetic variants that affect how abundant are different messenger forms.”
Professor Manolis Dermitzakis from the Genetics Department of the Faculty of medicine of the University of Geneva, formerly at the Sanger Institute
The results of this study have implications for human health. It is known that DNA variants affecting gene activity may be responsible for susceptibility to common diseases such as diabetes, cardiovascular diseases and asthma. The understanding of how the subtle differences uncovered in this study modulate gene expression should boost the understanding of their mechanisms at the cellular level, enabling faster and more focused development of treatments.
This study provides a framework towards the full understanding of the impact of genetic variations in cellular interactions, which has important implications for the understanding of human diseases.
The study was carried out by researchers from the Wellcome trust Sanger Institute in Cambridge, UK, the University of Geneva in Switzerland, and the Centre de Regulacio Genomica in Barcelona, Spain.
More information
Funding
This work was supported by the Wellcome Trust, Louis-Jeantet Foundation, Swiss NSF Frontiers in genetics, Spanish Ministry of Science and Consolider.
Participating Centres
- Department of Genetic Medicine and Development, University of Geneva Medical School, Geneva, Switzerland
- Wellcome Trust Sanger Institute, Cambridge, UK
- Center for Genomic Regulation, University Pompeu Fabra, Barcelona, Spain
Publications:
Selected websites
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute, which receives the majority of its funding from the Wellcome Trust, was founded in 1992. The Institute is responsible for the completion of the sequence of approximately one-third of the human genome as well as genomes of model organisms and more than 90 pathogen genomes. In October 2006, new funding was awarded by the Wellcome Trust to exploit the wealth of genome data now available to answer important questions about health and disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


