Complete fluke? Genome sequencers crack parasite genome
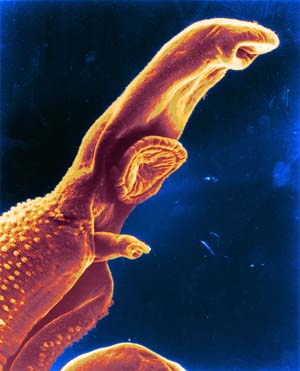
Researchers have today published the complete genome sequence of the Schistosoma mansoni, a parasitic worm – commonly known as a blood fluke – that causes devastating disease. The World Health Organization ranks schistosomiasis as a neglected disease of the poor, affecting 210 million people in 76 countries, and each year causing 280,000 deaths in sub-Saharan Africa alone.
The international team has identified several potential new drug targets and the genome sequence will be invaluable to scientists searching for new methods to treat and eradicate the disease.
Schistosomiasis has devastating global impact, yet research has been neglected for years. In part, this is due to the huge challenges that biologists face when studying the organism. Currently, there is only one drug treatment that is used to treat schistosomiasis and – with mounting fears that the parasites will become resistant – researchers have been looking at ways to find new drug targets. Today’s publication provides the first steps.
“This genome sequence catapults schistosomiasis research into a new era. It provides a foundation for understanding aspects of the parasite’s complex biology as well as a vehicle to immediately identify new targets for drug treatment.”
Dr Matthew Berriman of the Wellcome Trust Sanger Institute and first author and co-leader of the study
To identify new targets for drugs to combat pathogens Researchers most often exploit differences between the pathogen and its human host. However, S. mansoni is far closer to us in evolutionary terms than many of the major parasites whose genomes have been sequenced, such as malaria or trypanosomes. S. mansoni is a primitive animal, so the team could use a novel approach: they deliberately looked for similarities between humans and the parasite to try to exploit the activities of existing drugs, already on doctor’s shelves for other uses.
“This research provides the basis for scientists to discover the parasite’s weak spots and to exploit them for developing new drugs. As in past years, doctors rely on a single drug called praziquantel to treat patients infected with this parasite. We have already seen laboratory models of selection for resistance to drug treatments, and there have been some isolated reports of the parasite’s drug tolerance in countries affected by schistosomiasis.
“We need to take advantage of this genetic sequence data to find new and improved ways of coping with this problem that devastates much of the developing world.”
Michael Gottlieb, Associate Director of Science, Foundation for the National Institutes of Health
The researchers sequenced the parasite genome using the whole-genome shotgun method. They found that the genome – about ten times the size of the malaria parasite genome – contained almost 12,000 genes.
The team searched for possible novel drug targets. More than half of all known drugs are active against the proteins at the cell surface that act as gatekeepers or transmit signals. In their search, the teams focussed on these targets as well as processes that are key to the parasite’s survival, including lipid metabolism.
“Previous searches for treatments for schistosomiasis have involved in vivo screening using animal models. The genome sequence has given us, for the first time, a comprehensive view of the engines that drive the parasite, the strategies that allow it to survive in us, its human host. It is a catalogue of opportunities.”
Najib El-Sayed Associate Professor in the Department of Cell Biology and Molecular Genetics at the University of Maryland and co-leader on the study
The team used two approaches to find existing drugs that might be ‘recycled’ for use against schistosomes. A search of a database of current human drugs found matches to 26 genes in the parasite, a few of which are being explored, but many of which are novel.
The second approach was to ask: do parasite proteins resemble targets of available drugs? Matching parasite genes to known drug targets, the team found more than 90 candidates that meet minimal requirements of short treatment course and ready availability.
This work has to take account of the practicalities of drug distribution and searches for treatments must favour short-course, orally administered drugs. Existing therapies could be a rich source. Affordable treatments for schistosomiasis are vital: many affected countries are poverty stricken and lack robust healthcare systems.
As well as laying the foundations for improved treatment and perhaps eradication of schistosomiasis in the developing world, the team also learned more about the evolution of the simple species, exploring the beginnings of the evolution of animals.
“Blood flukes, such as S. mansoni, are flatworms that represent a poorly explored area of biology. Their genome sequence allows us to shed more light on the evolution of simple animals. Their body designs share a common plan with all animals from fish to humans.”
Dr Matthew Berriman of the Wellcome Trust Sanger Institute and first author and co-leader of the study
Like us, they develop from three major body layers and their bodies are ‘bilaterally symmetrical’ – structured with one line of symmetry only. Using the Treefam resource, hosted by the Sanger Institute, the team found gene functions that are either restricted to, or expanded in, more complex Bilateria species and absent from the more simple sea anemone: genes from the dawn of this important change in body structure.
S. mansoni is one of three species of the Schistosoma parasite that, because of a deficit in lipid levels, are dependent on a human host to complete their life cycle: they need our lipids to survive. Humans are infected through contact with contaminated water: the larvae of the parasite are released by aquatic snails and penetrate human skin.
The parasites develop in the blood system of the human and can also pass to the liver, lungs and bladder. When mature, they lay eggs in the intestinal wall: while some of the eggs are passed as faeces, surviving eggs can travel to the liver, where they induce in an extreme inflammatory immune response, which is the cause of the schistosomiasis.
The publication of the S. mansoni genome comes alongside that of the S.japonicum species, also a common cause of schistosomiasis. Together, the sequences provide new avenues for study in evolution, genetics and functional genomics.
Schistosomiasis is one of 14 Neglected Tropical Diseases listed by World Health Organization, who assert that 20 million people suffer with severe consequences from the disease: S. mansoni is prevalent in Africa, Mediterranean regions, the Caribbean and South America: S. aponicum is found in Eastern Asia.
The authors conclude that genome sequence can provide rich, fertile ground for exploration: “The sequence provides the scientific community with multiple avenues to study this under-researched human pathogen and will drive future evolutionary, genetic and functional genomic research. Not least, given that just one drug is widely available to treat schistosomiasis, the genome sequence, including the genome-mining analysis presented, offers the possibility that new drug candidates will be identified soon.”
More information
Funding
This research was funded by the Wellcome Trust and the National Institutes of Health-National Institute of Allergy and Infectious Diseases, the Oyama Health Foundation, Japan Society for the Promotion of Science, MEXT, The Sandler Foundation, FAPEMIG Brazil, the PhRMA Foundation, The Burroughs Wellcome Fund and The UNICEF/UNDP/World bank/WHO Special program for research and training in tropical diseases (TDR).
Participating Centres
- Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, UK
- Maryland Pathogen Research Institute, University of Maryland, College Park, MD, USA
- The Institute for Genomic Research/The J. Craig Venter Institute, 9712 Medical Center Dr., Rockville, MD, USA
- Departments of Biochemistry and Pathology, University of Texas, Health Science Center, San Antonio, TX, USA
- Department of Biology, University of York, York, UK
- Department of Cell Biology and Molecular Genetics, University of Maryland, College Park, MD, USA
- Center for Bioinformatics and Computational Biology, University of Maryland, College Park, MD, USA
- Sandler Center for Basic Research in Parasitic Diseases, California Institute for Quantitative Biomedical Research, Byers Hall, University of California, San Francisco, CA, USA
- Departments of Biopharmaceutical Sciences and Pharmaceutical Chemistry,California Institute for Quantitative Biomedical Research, Byers Hall, University of California, San Francisco, USA
- Cancer Research UK Centre for Cancer Therapeutics, The Institute of Cancer
- Research, Haddow Laboratories, Belmont, Sutton, Surrey, UK
- Centro de Pesquisas René Rachou – FIOCRU, Belo Horizonte, MG 30190002, Brazil
- Department of Microbiology, University College Cork, Ireland
- European Bioinformatics Institute (EMBL-EBI), Wellcome Trust Genome Campus, Hinxton, Cambridge, UK
- Department of Biomedical Sciences, Iowa State University, Ames, USA
- Instituto de Química and 16Instituto de Física de São Carlos, Universidade de São Paulo, Brazil
- Primate Research Institute, Kyoto University, Inuyama, Aichi, Japan
- Biomedical Parasitology Division, The Natural History Museum, London, UK
- Inserm, U 547, Université Lille, Institut Pasteur de Lille, Lille, France
- Institut für Mikrobiologie und Genetik, Abteilung Bioinformatik, Universität Göttingen, Göttingen, Germany
- Department of Biological Sciences, Illinois State University, Normal, IL, USA
Publications:
Selected websites
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute, which receives the majority of its funding from the Wellcome Trust, was founded in 1992. The Institute is responsible for the completion of the sequence of approximately one-third of the human genome as well as genomes of model organisms and more than 90 pathogen genomes. In October 2006, new funding was awarded by the Wellcome Trust to exploit the wealth of genome data now available to answer important questions about health and disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


