Switches, circuits, brain and disease
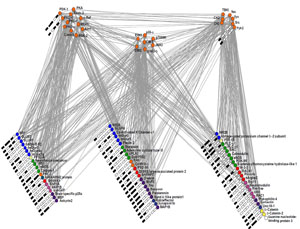
Scientists today announce a breakthrough that will lead to fundamental new understanding of the workings of the brain and its diseases. Research teams, led by Professor Seth Grant and Dr Jyoti Choudhary, from the Wellcome Trust Sanger Institute, have uncovered the first evidence that neurotransmitters control circuits of molecules inside synapses.
For over a century, mankind has known that the brain is the most complex of organs, with a million billion nerve cells connected to one another at specialised junctions – called synapses. These nerve cells communicate at synapses using chemicals – called neurotransmitters – that are released from a nerve cell to activate the connected nerve cell.
“We have uncovered a remarkable molecular circuitry that is no less impressive than the neuronal circuits of the brain. The molecular machinery in each synapse contains hundreds of proteins communicating with each other in the most intricate of networks.”
Professor Seth Grant Leader of the Genes to Cognition programme at the Wellcome Trust Sanger Institute
Using a combination of state-of-the-art proteomic and computational methods, the lead authors on the study – Dr Marcelo Coba, Dr Mark Collins and Dr Andrew Pocklington – mapped the details of the molecular networks. The team discovered that the molecular networks were structured much like computer chips inside a computer, containing many transistors and switches that can perform the calculations underlying the power of the brain.
The team’s discoveries led researchers to the conclusion that the brain is organised like the internet, where billions of these molecular computers – intricately complex in themselves – are connected by billions of nerve cells.
“The study by Coba and colleagues gives us a new insight into yet another layer of complexity in the brain. They combine network-modeling approaches to develop an elegant model that starts to unravel this complexity revealing a logical architecture. The new insights into the biochemical cascades that regulate information processing in the brain open up new opportunities for modeling how the brain works in normal as well as during disease.”
Professor Douglas Armstrong from the Institute for Adaptive and Neural Computation at the University of Edinburgh
These new insights have important implications for understanding how humans think: how we learn and how we remember.
For years it has been recognised that synapses can change with experience and it has been roundly acknowledged that this must be a mechanism for learning and memory.
However The existing molecular models have not been able to explain aspects of long-term memories, and this has led theoreticians to propose there must be a molecular mechanism yet to be discovered. These experts predicted that there was specialised molecular machinery with exactly the properties of the networks discovered in this study. These molecular circuits are ideally suited to improve the memory storage capacity of the brain.
Using the maps of the molecular circuits, the scientists investigated how these networks could either be disrupted in diseases or be used as therapeutic drug targets. Remarkably, they found that many of the proteins in the molecular networks were integral in brain diseases including depression, schizophrenia, autism, Alzheimer’s and Parkinson’s disease. These disease proteins were found to be connected to the neurotransmitter receptors, which are the site of action of many therapeutic drugs.
The molecular networks are part of a specialised set of proteins in the synapse: these include cadherin-10 (CDH10), which was shown in research published on 28 April 2009 to be a risk gene in autism.
“Defining these molecular circuits will be invaluable for the development of therapeutics for a range of neuropsychiatric disorders. This is a major goal of pharmaceutical companies.”
Professor Seth Grant, Sanger Institute
The new findings build on recent discoveries by Professor Grant and his colleagues that found that these molecular networks have evolved from simple animals and have become increasingly intricate with the development of more complex species, including humans.
More information
Funding
This work was funded by the Wellcome Trust
Collaborating Institutions
- Wellcome Trust Sanger Institute, Hinxton, Cambridgeshire, UK
- Institute for Adaptive and Neural Computation, Division of Informatics, University of Edinburgh, Edinburgh, UK
Publications:
Selected websites
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute, which receives the majority of its funding from the Wellcome Trust, was founded in 1992. The Institute is responsible for the completion of the sequence of approximately one-third of the human genome as well as genomes of model organisms and more than 90 pathogen genomes. In October 2006, new funding was awarded by the Wellcome Trust to exploit the wealth of genome data now available to answer important questions about health and disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


