Simple yeast leads to understanding ourselves
A new study uncovers the evolutionary history of the organism that gives us fermented beer and risen bread. Using sequencing technology researchers have, for the first time, shown that humans have tamed yeast on multiple occasions in their history.
More dramatically, the research links two important projects in biomedical research and in the work of the Wellcome Trust Sanger Institute. Yeast was the first genome completed at the Sanger Institute in 1996 and the research published today has played an essential role in developing the principles and tools for The 1000 Genomes Project launched in 2007.
From cell division to cancer, yeast has helped researchers to grasp biological principles and today’s results continue that tradition. But has our long association with yeast made it too domesticated? Does the highly selected yeast we now produce for laboratory experiments in any way influence the outcome of scientific research?
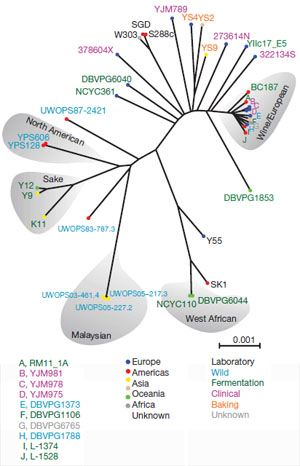
The new research looked at the genome sequences of 70 samples of yeast from around the world.
“We didn’t know much about the evolution of yeast and how much influence we had over the centuries by highly selecting for brewing, baking and laboratory experiment properties. It remains to this day one of the best models for evolutionary studies.”
Ed Louis A professor of genome dynamics at the Institute of Genetics at The University of Nottingham
Until now little has been known about the evolutionary processes taking place within populations and different strains of domesticated yeast. The team examined genome sequences from Saccharomyces cerevisiae (S. cerevisiae), commonly used in laboratory research, baking, wine making and in the production of sake. These were compared with its closest relative, Saccharomyces paradoxus (S. paradoxus) isolates, found in oak tree bark.
The project did not sequence the genome of each strain in great detail: to improve efficiency, each was sequenced to a depth that would allow the team to gather DNA sequence differences between the strains and, where sequence was poor or lacking, to impute the probable sequence differences between them. The techniques and the results have served as proof of principle for the methods used in The 1000 Genome Project, the three-year programme to decode genomes of 1000 people.
“We have been able to use the amazing power of modern DNA sequencing to obtain an unprecedented picture of the way the genes in this organism vary, and how it evolved. The tools we developed to study yeast are now being applied to the human genome, to help identify genetic variants that are involved in disease.”
Dr Richard Durbin Senior Investigator at the Wellcome Trust Sanger Institute and co-leader of The 1000 Genomes Project
The genome of S. cerevisiae – it is about 12 million letters of genetic code – was sequenced by an international consortium including the Wellcome Trust Sanger Institute – the process took years to complete, with the research being published in 1996. Using the next-generation sequencing technologies now available, researchers can complete a sequence of this size and quality in little over half an hour.
“Model organisms such as yeast, worm, fly and mouse have been and will continue to be essential to biological exploration.”
Dr Durbin whose own work was instrumental in interpreting the genome of the nematode worm and in the Human Genome Project
The use of yeasts in cooking and brewing has led to suggestions that humans might have cultivated selected strains for these purposes. In at least some cases, nature still has a lead. The research team mention recent findings, in Malaysia, of chronic intake of alcoholic nectar from Bertram palms by wild tree shrews: fermented beverages seem popular with other primates and not only humans.
More information
Funding
This work was funded by the Wellcome Trust, Royal Society, Biotechnology and Biological Sciences Research Council, Swedish Research Council, Swedish Foundation for Strategic Research, National Environment Research Council, Canada Foundation for Innovation, National Science Foundation, National Institutes of Health and a Hertz fellowship.
Participating Centres
- Institute of Genetics, Queen’s Medical Centre, University of Nottingham, Nottingham, UK
- Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK
- Department of Cell & Systems Biology, University of Toronto, Ontario, Canada
- Department of Cell and Molecular Biology, Lundberg Laboratory, University of Gothenburg, Medicinaregatan 9c, Gothenburg, Sweden
- National Collection of Yeast Cultures, Institute of Food Research, Norwich Research Park, Colney, Norwich, UK
- Division of Biology, Imperial College London, Silwood Park, Ascot, UK
- Faculty of Life Sciences, University of Manchester, Manchester, UK
- Department of Physics, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA
Publications:
Selected websites
The University of Nottingham
The University of Nottingham is ranked in the UK’s Top 10 and the World’s Top 100 universities by the Shanghai Jiao Tong (SJTU) and Times Higher (THE) World University Rankings.
More than 90 per cent of research at The University of Nottingham is of international quality, according to RAE 2008, with almost 60 per cent of all research defined as ‘world-leading’ or ‘internationally excellent’. Research Fortnight analysis of RAE 2008 ranks the University 7th in the UK by research power. In 27 subject areas, the University features in the UK Top Ten, with 14 of those in the Top Five.
The University provides innovative and top quality teaching, undertakes world-changing research, and attracts talented staff and students from 150 nations. Described by The Times as Britain’s “only truly global university”, it has invested continuously in award-winning campuses in the United Kingdom, China and Malaysia. Twice since 2003 its research and teaching academics have won Nobel Prizes. The University has won the Queen’s Award for Enterprise in both 2006 (International Trade) and 2007 (Innovation – School of Pharmacy), and was named ‘Entrepreneurial University of the Year’ at the Times Higher Education Awards 2008.
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute, which receives the majority of its funding from the Wellcome Trust, was founded in 1992. The Institute is responsible for the completion of the sequence of approximately one-third of the human genome as well as genomes of model organisms and more than 90 pathogen genomes. In October 2006, new funding was awarded by the Wellcome Trust to exploit the wealth of genome data now available to answer important questions about health and disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


