Maps of Structural Variation released for Cancer Cell Lines
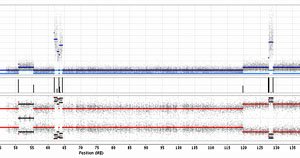
The Wellcome Trust Sanger Institute announces today the release of a new dataset cataloguing the structural genomic changes in almost 800 cancer cell lines. The Cancer Genome Project has recently completed the genome-wide analysis of copy number changes in a Core Set of cancer cell lines. Cancer cell lines are invaluable reagents in investigating the molecular biology of the disease as well as important tools in developing new therapeutic approaches.
This core set of cancer cell lines includes samples from most types of human cancer; the study also included a series of over 460 normal control samples. In total nearly 1250 samples were analysed using the Affymetrix SNP6.0 Whole Genome Array, which evaluates 1.8 million points in the genome simultaneously, averaging one data point every 1700 base-pairs of the 3 billion base-pairs that make up the human genome. In all, over 2 billion data points were captured and subjected to analysis using newly developed algorithms specifically designed to analyse allele-specific copy number changes in complex cancer genomes.
“Development of a new in-house pipeline used hidden Markov models to extract maximum information from the data. This accurately identified genomic regions of fixed allelic copy number, helping pinpoint causative changes in cancer.”
Dr Chris Greenman of the Cancer Genome Project and the scientist in charge of the data analysis
Using this information the team from the Wellcome Trust Sanger Institute were able to produce genomic maps for each sample, highlighting the alterations that have occurred during the genesis of the cancer. These structural alterations can inactivate genes, produce multiple copies of genes thereby increasing gene activity or, more rarely, result in fusion of two genes, leading to gene products possessing new properties, for instance fusion of the BCR and ABL1 genes in chronic myeloid leukaemia.
“Previously, using an earlier version of the current array, we developed maps of structural variation at lower resolution. The new dataset provides a much more refined view of the changes that have occurred in these samples as it represents a 180-fold increase in resolution. When viewed across nearly 800 samples these maps give a remarkable insight into the ravages of the genome induced during cancer development.”
Dr Graham Bignell Project Leader at the Wellcome Trust Sanger Institute
The data release includes a data-mining tool that extracts summary information across the sample set for both user-defined genomic regions and predefined loci. The summary information for known genes and the genomic maps for individual samples are freely available via COSMIC, together with the complete dataset broken down by sample, chromosome or tissue type via the CGP website. The segmentation tool used in these analyses is also being released. Higher resolution maps combined with a viewing tool are available together with the raw output files. These can be obtained from the CGP data archive subject to a data transfer agreement.
As part of the ongoing study within this series of cancer cell lines, mutations in more than 60 known cancer genes are being characterised, with all data being made publicly available on the COSMIC website. The data currently released provides an insight into the genomic alterations that have occurred within these samples as they progress towards cancer. Already the combination of these different datasets makes this the best characterised series of cancer samples available to the research community, this will be further enhanced as the project continues, adding to the utility of this resource.
Using a range of high-throughput approaches, the Sanger Institutes Cancer Genome Project (CGP), led by Professor Mike Stratton and Dr Andy Futreal, is focused on the characterisation of cancer genomes with the goal of identifying key genes that are important in the genesis of cancer. Such insights will ultimately allow for improved diagnosis and treatment of cancer.
Ultimately all cancers derive from changes to DNA. These changes are characterised by alterations which extend from single base changes to vastly complex changes involving large-scale gains and losses in DNA content. New technologies allow researchers to begin to catalogue and investigate these large-scale copy number changes in unprecedented detail.
More information
Selected websites
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute, which receives the majority of its funding from the Wellcome Trust, was founded in 1992. The Institute is responsible for the completion of the sequence of approximately one-third of the human genome as well as genomes of model organisms and more than 90 pathogen genomes. In October 2006, new funding was awarded by the Wellcome Trust to exploit the wealth of genome data now available to answer important questions about health and disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


