Crusader Y chromosomes in Lebanon
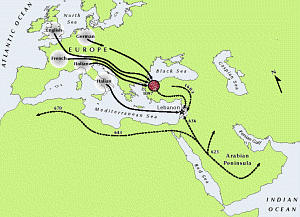
“We are fortunate that the history of Lebanon is so well documented. History gives us a perfect starting point and historians have known about these migrations for centuries. Now we geneticists can detect them as well. It shows how powerful genetics has become and is great news for future studies of places where we don’t know so much about the history.”
Genographic Associate Researcher Chris Tyler-Smith At the Wellcome Trust Sanger Institute
The analysis of the Y chromosome, a “genetic surname” specific to men, reveals that Lebanese populations are very closely related, and the novel findings result from a detailed analysis of many more markers and many more people than has been attempted before. However, over the past 1400 years geographically distinct populations from Europe and the Arabian Peninsula have entered Lebanon.
The data revealed several interesting findings. A genetic signature called WES1 that is found only in European populations was found also in Lebanese Christian men. Also, Lebanese Muslim men have very high frequencies of J1, typical of the populations of the Arabian Peninsula that were involved in the Muslim expansion. The study found no impact of the Ottoman expansion from Turkey in the 16th century.
This is the most careful and comprehensive study of these populations ever undertaken, and it’s revealed new insights into the complex history of my country. The subtle effects we have detected are really exciting but should not obscure the extensive underlying similarity among all of the populations of present-day Lebanon, a country that for too long has been divided along religious lines.”
Pierre Zalloua Genographic Principal Investigator, Middle East/North Africa
The study made use of comparative data from Genographic public participants living in Europe. Over 8,000 people from France, Italy and the UK have now participated – anonymously – in the project, and a subset of their data was used for comparative purposes to track the expansion of these lineages during the time of the Crusades.
“The massive database that we have assembled from Middle Eastern and European populations, including Genographic public participants, has allowed us to detect the subtle genetic impact of these historical events. This study illustrates the power of large amounts of data, and demonstrates how public participants from around the world can help to decipher historical migratory events.”
Dr Spencer Wells Genographic Project Director
More information
Funding
This project was supported in part by a grant from the National Geographic Committee for Research and Exploration; YX and CTS were supported by the Wellcome Trust. We thank Janet Ziegle and Applied Biosystems for providing STR genotyping and QA support. The Genographic Project is supported by funding from the National Geographic Society, IBM, and the Waitt Family Foundation.
Publications:
Selected websites
The Genographic Project
The Genographic Project was launched in 2005, by National Geographic and IBM, with field research supported by the Waitt Family Foundation, using genetics as a tool to address anthropological questions on a global scale. At the core of the project is a global consortium of ten regional scientific teams following an ethical and scientific framework and who are responsible for sample collection and analysis in their respective regions. The Project is open to members of the public to participate through purchasing a public participation kit from the Genographic website, where they can also choose to donate their genetic results to the expanding database. Sales of the kits are returned to the research and a Legacy Fund for indigenous peoples’ community-led language revitalization and cultural projects.
National Geographic
Founded in 1888, National Geographic is one of the world’s largest nonprofit scientific and educational organizations . Its mission is to increase and diffuse geographic knowledge while inspiring people to care about the planet. National Geographic reflects the world through its six magazines, cable television channels and programs, films, radio, books, videos, maps, interactive media and merchandise, reaching as many as 300 million people each month.
IBM
IBM is the world’s largest information technology company, with more than 80 years of leadership in helping businesses innovate. It has a long history of innovating on behalf of society, and in recent years has launched a series of major research initiatives designed to overcome many of the remaining "grand challenges" of science, including the Deep Blue chess-playing computer and unraveling the mysteries of protein folding with BlueGene, the world’s fastest supercomputer. IBM Research is the world’s largest information technology research organization , with more than 3,000 scientists and engineers at eight labs in six countries.
About the Waitt Family Foundation
Established in 1993 by Gateway Computer founder and now Chairman Ted Waitt and his wife, Joan, the Waitt Family Foundation focuses on humankind’s past, present and future. Specifically, the foundation funds projects aimed at making discoveries about our past that will help inform the way we are today and reveal untapped possibilities for the future.
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute, which receives the majority of its funding from the Wellcome Trust, was founded in 1992. The Institute is responsible for the completion of the sequence of approximately one-third of the human genome as well as genomes of model organisms and more than 90 pathogen genomes. In October 2006, new funding was awarded by the Wellcome Trust to exploit the wealth of genome data now available to answer important questions about health and disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


