Potent possibilities for parasite attack
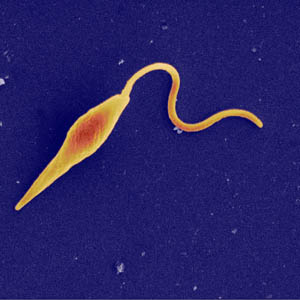
A comparison of three parasite species that cause Leishmaniasis has identified a small number of genes, many new to biology, that will provide a framework to target the search for new treatments. Leishmaniasis is a devastating disease that affects about two million people each year and threatens one-fifth of the world’s population and new treatments are desperately needed.
In their report in Nature Genetics, published online on Sunday 17 June 2007, the researchers compared the genomes of L. infantum and L. braziliensis, which cause life-threatening visceral and disfiguring mucocutaneous leishmaniasis, respectively, with the sequence they produced in 2005 for L. major, which causes a less severe, cutaneous form of the disease. Despite the major differences in disease type, only 200 out of more than 8000 genes present in each genome were found to be differentially distributed between the three species. This exceptionally small variation in gene content has given new insights into those processes that may determine disease severity in humans.
“Identifying factors that allow three closely related organisms to cause vastly different clinical outcomes is a major quest for researchers and in this study we have narrowed the search to a number that can be realistically studied.”
Dr Matt Berriman, senior author on the paper, from the Wellcome Trust Sanger Institute
The researchers found only five genes in the L. major genome for which no trace could be found in the other two species. By contrast, in Plasmodium, which causes malaria, about 20% of genes differ between related species.
“Clearly there must have been considerable evolutionary pressure over time to maintain the structure and sequence of the Leishmania genomes – the degree of similarity between these species was unexpected. Perhaps only a few parasite genes are important in determining which type of disease develops after infection and the host genome plays a major role in clinical outcome.”
Professor Deborah Smith, collaborator on this project at the University of York
The results picked up another surprising finding: the team could assign a function to only one-third of the 200 genes restricted to one or two of the species.
“The genome sequences have given us a short-cut to a small number of largely novel genes. Given their lack of similarity to human genes, they present a limited repertoire of potential targets for drug and vaccine development allowing researchers to optimise the use of limited resources.”
Dr Chris Peacock, first author on the report
Leishmaniasis is one of the neglected diseases that desperately need new research, as WHO/TDR notes: “Treatment of visceral leishmanisis by first-line drugs is long (4 weeks), given systemically, and expensive (US$120 – 150).” The affordable drugs have been in use for more than half a century and drug resistance is rife, creating a desperate need for new treatments. Biological studies for the function of 50 per cent of Leishmania genes are lacking, so this comparative genome study provides a route to find those that might be essential to each species.
One potential target is the CFAS gene that codes for cyclopropane fatty acid synthase, an enzyme that may be involved in producing components of the cell membrane. CFAS is present in the genomes of L. braziliensis and L. infantum, but is absent from the human genome. The parasite genes are thought to have been acquired from bacterial species that have very similar sequences.
“CFAS is involved in virulence and persistence in Mycobacterium, causative agent of tuberculosis, so the identification of a CFAS gene in Leishmania raises the exciting possibility that some virulence factors are conserved between bacterial and eukaryotic intracellular pathogens.”
Jeremy Mottram, a collaborator on the project who is a Professor in the Wellcome Centre for Molecular Parasitology at the University of Glasgow
Some families of genes that determine the properties of the parasite cell surface have grown in number and some declined among the three species: ‘death’ of genes seems to be a major force for differences between the parasite genomes. Some genes, however, are evolving rapidly, leading the team to suspect they include key genes involved in interacting with the human host – where the battle between parasite and patient is fought and where rapid response is important to both.
Remarkably, L. braziliensis, the most ancient Leishmania species sequenced, contains genes that could provide a working pathway for RNAi, an emerging mechanism for gene regulation. The genome sequences show that components for this pathway are absent from the other two Leishmania species. This pathway might serve as an experimental tool in understanding the role of the many genes whose function is unknown, by using experimentally induced RNAi to ‘knock-down’ gene activity prior to host infection.
“In addition to their function with respect to promoting diverse clinical outcomes, the remarkably limited number of species-specific genes should lead to the more rapid identification of sequences involved in specialized aspects of Leishmania biology, such as the development of L. braziliensis in the hindgut of its sandfly vector, and the restricted reservoir host range seen with L. infantum infections in dogs.”
David Sacks, PhD, Head of the Intracellular Parasite Biology Section at the National Institute of Allergy and Infectious Diseases, Bethesda, USA
Around 350 million people in 88 countries on four continents are at risk of Leishmaniasis and its incidence has risen sharply over the past 10 years. It is transmitted by the bite of various species of sandfly: wild and domesticated animals – as well as humans – act as a reservoir for the disease.
More information
Participating Centres
- Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK
- Immunology and Infection Unit, Department of Biology, University of York, and The Hull York Medical School, UK
- Wellcome Centre for Molecular Parasitology and Division of Infection & Immunity, Glasgow Biomedical Research Centre, University of Glasgow, 120 University Place, Glasgow, UK
- Departmento de Biologia Celular e Molecular e Bioagentes Patogênicos, Faculdade de Medicina, de Ribeirão Preto, Universidade de São Paulo, Av. Bandeirantes, 3900, Ribeirão Preto, São Paulo, Brazil
- Laboratoire de Génomique Fonctionnelle des Trypanosomatides, Université Victoir Segalen Bordeaux II, UMR-5162 CNRS, Bordeaux, France
Websites
- WHO Research and Training in Tropical Diseases (TDR) pages – http://www.who.int/tdr/diseases/leish/
- WHO Leishmaniasis pages – http://www.who.int/leishmaniasis/en/
- Leishmania major publication in Science – http://www.sciencemag.org/cgi/content/abstract/309/5733/436
- Immunology and Infection Unit, York – http://www.york.ac.uk/res/iiu/
- Wellcome Centre for Molecular Parasitology and Division of Infection & Immunity, University of Glasgow – http://www.gla.ac.uk/centres/wcmp/
- Departmento de Biologia Celular e Molecular e Bioagentes Patogênicos, Faculdade de Medicina, de Ribeirão Preto, Universidade de São Paulo, Sao Paulo – http://rbp.fmrp.usp.br/
- Université Victor Segalen Bordeaux 2 – http://www.u-bordeaux2.fr/
Funding
This study was funded by the Wellcome Trust through its support of the Pathogen Sequencing Unit at the Wellcome Trust Sanger Institute. Individual researchers were supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) fellowships, a postgraduate studentship from the Biotechnology and Biological Sciences Research Council and support from the UNICEF/UNDP/WORLD BANK/WHO Special Programme for Research and Training in Tropical Diseases (TDR).
Publications:
Selected websites
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute, which receives the majority of its funding from the Wellcome Trust, was founded in 1992. The Institute is responsible for the completion of the sequence of approximately one-third of the human genome as well as genomes of model organisms and more than 90 pathogen genomes. In October 2006, new funding was awarded by the Wellcome Trust to exploit the wealth of genome data now available to answer important questions about health and disease.
The Wellcome Trust
The Wellcome Trust is the largest charity in the UK. It funds innovative biomedical research, in the UK and internationally, spending around £500 million each year to support the brightest scientists with the best ideas. The Wellcome Trust supports public debate about biomedical research and its impact on health and wellbeing.


