When Less is More
 Some genes are quite simply bad for us, researchers at the Wellcome Trust Sanger Institute suggest today. They asked the question: what if our behaviour has changed such that genes that suited us in our past no longer benefited us? Could our genome keep up?
Some genes are quite simply bad for us, researchers at the Wellcome Trust Sanger Institute suggest today. They asked the question: what if our behaviour has changed such that genes that suited us in our past no longer benefited us? Could our genome keep up?
Their remarkable study, published in the April edition of the American Journal of Human Genetics, suggests that a gene called caspase-12 has been inactivated in the human population because the active gene can lead to poorer response to bacterial infection. When infectious diseases became more common in human populations, perhaps because population densities grew and pathogens were able to spread more rapidly, the people with the inactive gene were at an advantage and prospered.
The wheels of evolution grind exceedingly slow and our genetic make up today reflects the forces that shaped our genome since our divergence from chimpanzees perhaps six million years ago and beyond. Modern human anatomy appeared almost 200,000 years ago (200 KYA) but modern behaviours emerged much later, around the time of migration from Africa, perhaps 100-50 KYA.
Genes that are important in our development as a species help both in understanding human history and behaviour and in understanding human disease. One of the best-known examples of the advantage of losing a functioning gene is the variant CCR5 ?32, which renders people who carry it much more resistant to infection with HIV and to development of AIDS.
If circumstances arise where loss of activity of an existing gene confers an advantage, then there are always likely to be a few individuals with inactive genes in a population, and these can then spread. Caspase-12 is an example of such a gene inactivation.
“Caspase-12 works as a part of the system that responds to bacterial infection and people with the fully working gene produce lower levels of response. However, if bacteria enter the bloodstream, these people are at a greater danger of bacterial sepsis.”
Dr Chris Tyler-Smith Principal Investigator at the Wellcome Trust Sanger Institute
Sepsis – the growth of bacteria in the bloodstream – is the most common cause of death in infants and children in the world and deaths ascribed to the four major killers (pneumonia, diarrhoea, malaria and measles) often occur via a common pathway leading to fatal sepsis.
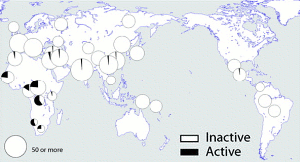
“The protein is active in perhaps 20 per cent of individuals in Africa, but the active variant is rare in the rest of the worlds population. In this study we asked the questions: How and when did the inactive form become predominant?”
Dr Tyler-Smith Sanger Institute
Using samples from the HGDP-CEPH and HapMap collections of DNAs, the team first catalogued the relative frequency of the active and inactive forms of caspase-12 in over 1000 individuals and sequenced the gene in 77. They found no examples of the active form in most populations and only rare examples (<1 per cent) in samples from Israel, China, Pakistan and Mexico; by contrast, the active version was prevalent in African populations (28 per cent of individuals from sub-Saharan Africa) and predominated in two groups.
The amounts of DNA sequence variation in the chromosomes bearing active genes were much higher than in chromosomes bearing inactive genes: inactive genes were carried on more uniform chromosome backgrounds. This suggests that either the population passed through a ‘bottleneck’ (giving reduced variation in a small population) or the inactive form was actively favoured and selected in our recent past.
To distinguish these, Dr Tyler-Smith and his colleagues looked at blocks of variation called haplotypes in the region of the caspase gene. All eight haplotypes for the active gene differed from one another; by contrast, almost two-thirds of the haplotypes for the inactive gene were identical. High similarity or identity of haplotypes is a feature of recent positive selection – chromosomes originating from one individual confer an advantage and spread through succeeding generations.
From studying the similarities and differences, the team suggest that the inactivating mutation occurred about 400-550 KYA. But when did these mutations spread through the population and what could have favoured their spread?
“The mutation seems to have spread just before the migration out of Africa. What we see is not a bottleneck, but changes in the populations that favour the inactive gene.”
Dr Chris Tyler-Smith Sanger Institute
Survival of severe sepsis would have been a strong force for selection of the inactive gene, which would have been greatly favoured, especially before improved sanitation. Today, people with two copies of the inactive gene are eightfold more likely to escape severe sepsis and threefold more likely to survive.
But the study leaves us with a puzzle. If it is so good to have the inactive gene, why did our ancestors have an active form in the first place? Probably the active gene is better in some circumstances, and the inactive gene in others. In most of the world today, it is advantageous to have the inactive gene, but this need not always be so.
“All organisms are dynamic and are fitted to the environment as it existed in the past. We should understand that our genetic constitution is in flux and we will continue to adapt. No genome is perfect and none is ideally suited to all environments. People sometimes ask whether humans are still evolving. Clearly, we are.”
Dr Chris Tyler-Smith Sanger Institute
More information
Participating Centres
- The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, UK
- Human Genetics Department, University of Chicago, Chicago, IL 60637, USA
- Department of Biology, University of Rochester, Rochester, NY 14627, USA
- Broad Institute of the Massachusetts Institute of Technology, and Harvard University, Cambridge, Massachusetts, USA
- European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, UK
Publications:
Selected websites
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute, which receives the majority of its funding from the Wellcome Trust, was founded in 1992. The Institute is responsible for the completion of the sequence of approximately one-third of the human genome as well as genomes of model organisms and more than 90 pathogen genomes. In October 2006, new funding was awarded by the Wellcome Trust to exploit the wealth of genome data now available to answer important questions about health and disease.
The Wellcome Trust and Its Founder
The Wellcome Trust is the most diverse biomedical research charity in the world, spending about £450 million every year both in the UK and internationally to support and promote research that will improve the health of humans and animals. The Trust was established under the will of Sir Henry Wellcome, and is funded from a private endowment, which is managed with long-term stability and growth in mind.


