New insights into Chlamydial Infection
Today, the hiding places for that killer – a bacterium called Chlamydophila abortus – are fewer because of the genome sequence produced in a collaboration between the Wellcome Trust Sanger Institute, the Moredun Research Institute and the Scottish Crop Research Institute.
Infection with Chlamydophila abortus is the most common cause of infectious abortion in sheep in the UK, leading to loss of lambs and economic costs of around £30M each year. Chlamydial infections of humans and animals are of enormous public and animal health significance worldwide, both in terms of disease and economic impact.
The genome sequence, published in Genome Research, will bring new possibilities in the fight to control the spread of infection.
“Genomics has transformed this field because we had a complete lack of genetic tools for research directed at combating chlamydial disease. Comparing this genome with those of other chlamydial species sequenced to date and others that are in progress will help to identify proteins for vaccine development and for the development of specific diagnostic tests.”
Dr David Longbottom Head of Molecular Chlamydia Research at the Moredun Research Institute
Infection in domestic animals is thought to be through ingestion of bacteria deposited on the grass via infected placentas after birth. Although there are commercially available vaccines, the disease persists because of difficulties in diagnosis, in vaccine uptake and in management of the sheep flocks. Early diagnosis is important and the infection responds well to early treatment with antibiotics, particularly tetracyclines and erythromycin. The newly produced sequence will help in the identification of suitable targets for improving diagnosis and will aid vaccine development.
“The genome is very similar to the genome of the related organisms C. pneumoniae and C. caviae. However, we have identified variable families of proteins which will be key genes to study in the disease process and for intervention.”
Dr Nick Thomson Project Leader at the Wellcome Trust Sanger Institute
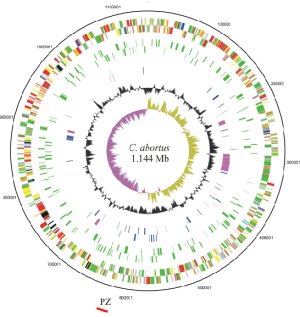
The genome paper describes the identification and characterisation of these proteins, which will form the basis for future research. The finished and annotated sequence consists of 1,144,377 base-pairs and codes for a predicted 961 proteins. The low number of genes suggests that the genome of C. abortus has been modified for a specific niche: as an intracellular infection, it can lay dormant between outbreaks and cause abortion of lambs in a following season. Individual animals carry the infection asymptomatically.
Especially with the coming of spring and lambing season, C. abortus also becomes a potential threat to human health: pregnant women exposed to infected animals are at possible risk of abortion and life-threatening illness. Most farms advise pregnant women not to come in contact with pregnant sheep or their lambs.
“We expect to see improvements in the development of new vaccines and diagnostic tests. Together with combined changes in flock management practices, these will ultimately reduce the environmental spread of infection to other animals and reduce the risks of zoonotic transmission to humans.”
Dr David Longbottom Head of Molecular Chlamydia Research at the Moredun Research Institute
Chlamydophila abortus is part of a family of bacteria that includes Chlamydia trachomatis, the most common sexually transmitted pathogen in the United Kingdom. It is an obligate intracellular organism – one that can divide only within a host cell. Inside the host cell, the bacteria develop in a specialized ‘compartment’ called an inclusion. This makes these organisms very difficult to study in the laboratory. The information gained from the genome sequence combined with that from the other chlamydial genomes will ultimately enable advances in research to address key questions on how these organisms cause disease.
More information
Funding
This two-year collaborative project was funded by the Scottish Executive Environment and Rural Affairs Department (SEERAD) and ended in March 2004. The Pathogen Sequencing Unit and Pfam Group are supported by the Wellcome Trust.
Participating Centres
- The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, CB10 1SA, UK
- Moredun Research Institute, Penicuik, Midlothian EH26 0PZ, UK
- Scottish Crop Research Institute, Invergowrie, Dundee DD2 5DA, UK
Publications:
Selected websites
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute, which receives the majority of its funding from the Wellcome Trust, was founded in 1992. The Institute is responsible for the completion of the sequence of approximately one-third of the human genome as well as genomes of model organisms and more than 90 pathogen genomes. In October 2006, new funding was awarded by the Wellcome Trust to exploit the wealth of genome data now available to answer important questions about health and disease.
The Wellcome Trust and Its Founder
The Wellcome Trust is the most diverse biomedical research charity in the world, spending about £450 million every year both in the UK and internationally to support and promote research that will improve the health of humans and animals. The Trust was established under the will of Sir Henry Wellcome, and is funded from a private endowment, which is managed with long-term stability and growth in mind.


